从空间解析的转录组学数据中绘制细胞相互作用图。
IF 36.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
细胞-细胞通讯(CCC)对生命的形成和功能至关重要。然而,准确、高通量地绘制一个细胞中所有基因的表达如何影响另一个细胞中所有基因表达的图谱,直到最近才通过引入空间分辨转录组学(SRT)技术,特别是那些实现单细胞分辨的技术而成为可能。然而,要正确分析这种高度复杂的数据仍面临巨大挑战。在这里,我们引入了一个多实例学习框架 Spacia,通过独特地利用 SRT 的空间模式,从 SRT 生成的数据中检测 CCC。我们强调了 Spacia 在克服用于推断 CCC 的流行分析工具的基本局限性方面的能力,这些局限性包括失去单细胞分辨率、仅限于配体-受体关系和先前的相互作用数据库、假阳性率高,以及最重要的一点,即缺乏对多发送方到单接收方范例的考虑。我们评估了 Spacia 对三种商业化单细胞分辨率 SRT 技术的适用性:MERSCOPE/Vizgen、CosMx/NanoString 和 Xenium/10x。总之,Spacia 是推进蜂窝通信定量理论的重要一步。本文章由计算机程序翻译,如有差异,请以英文原文为准。
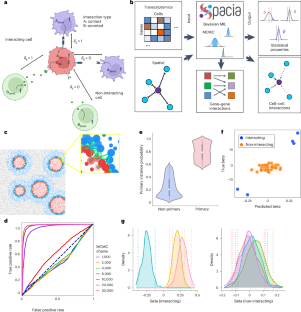
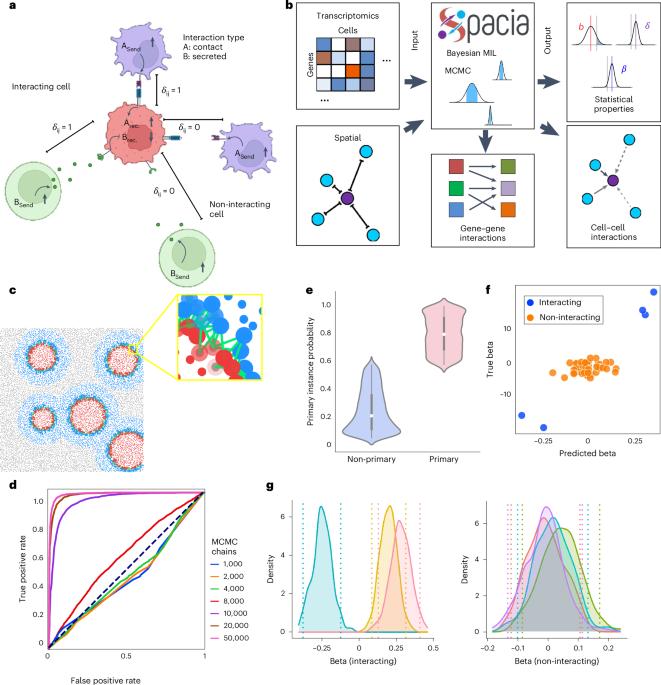
Mapping cellular interactions from spatially resolved transcriptomics data
Cell–cell communication (CCC) is essential to how life forms and functions. However, accurate, high-throughput mapping of how expression of all genes in one cell affects expression of all genes in another cell is made possible only recently through the introduction of spatially resolved transcriptomics (SRT) technologies, especially those that achieve single-cell resolution. Nevertheless, substantial challenges remain to analyze such highly complex data properly. Here, we introduce a multiple-instance learning framework, Spacia, to detect CCCs from data generated by SRTs, by uniquely exploiting their spatial modality. We highlight Spacia’s power to overcome fundamental limitations of popular analytical tools for inference of CCCs, including losing single-cell resolution, limited to ligand–receptor relationships and prior interaction databases, high false positive rates and, most importantly, the lack of consideration of the multiple-sender-to-one-receiver paradigm. We evaluated the fitness of Spacia for three commercialized single-cell resolution SRT technologies: MERSCOPE/Vizgen, CosMx/NanoString and Xenium/10x. Overall, Spacia represents a notable step in advancing quantitative theories of cellular communications. Spacia is a multiple-instance learning model for cell–cell communication (CCC) interference in single-cell resolution spatially resolved transcriptomics data. Spacia can map complex CCCs by modeling cell proximity and CCC-driven gene perturbation.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Methods
生物-生化研究方法
CiteScore
58.70
自引率
1.70%
发文量
326
审稿时长
1 months
期刊介绍:
Nature Methods is a monthly journal that focuses on publishing innovative methods and substantial enhancements to fundamental life sciences research techniques. Geared towards a diverse, interdisciplinary readership of researchers in academia and industry engaged in laboratory work, the journal offers new tools for research and emphasizes the immediate practical significance of the featured work. It publishes primary research papers and reviews recent technical and methodological advancements, with a particular interest in primary methods papers relevant to the biological and biomedical sciences. This includes methods rooted in chemistry with practical applications for studying biological problems.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


