癌症患者组织样本和细胞系的蛋白质表达图谱有助于深入了解肿瘤的异质性和依赖性。
IF 23.5
1区 医学
Q1 ONCOLOGY
引用次数: 0
摘要
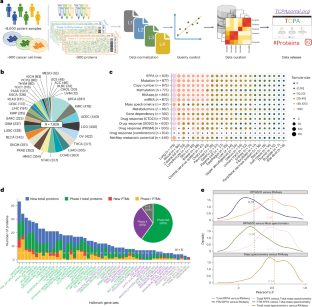
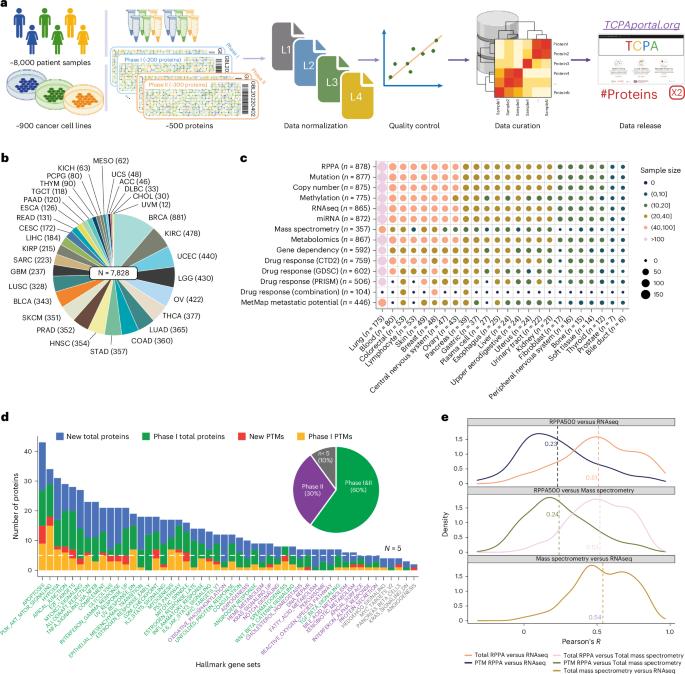
癌症基因组图谱(TCGA)和癌症细胞系百科全书(CCLE)是癌症研究的基础资源,提供了大量的分子和表型数据。然而,这些队列中不同癌症类型的大规模蛋白质组数据仍然有限。在此,我们在先前工作的基础上,利用反相蛋白质阵列生成了约 8,000 份 TCGA 患者样本和约 900 份 CCLE 细胞系样本的高质量蛋白质表达数据,涵盖 447 种临床相关蛋白质。这些蛋白质表达谱为了解肿瘤间异质性和癌症依赖性提供了深刻的见解,并可作为体细胞改变的灵敏功能读数。我们开发了一种以蛋白质为中心的系统化策略,用于鉴定合成致死对,并通过实验验证了蛋白激酶 A 亚基 α 与表皮生长因子受体之间的相互作用。我们还鉴定了具有临床意义的转移相关蛋白标记物。这个数据集是我们了解癌症机制、发现蛋白质生物标记物和开发创新治疗策略的宝贵资源。本文章由计算机程序翻译,如有差异,请以英文原文为准。
A protein expression atlas on tissue samples and cell lines from cancer patients provides insights into tumor heterogeneity and dependencies
The Cancer Genome Atlas (TCGA) and the Cancer Cell Line Encyclopedia (CCLE) are foundational resources in cancer research, providing extensive molecular and phenotypic data. However, large-scale proteomic data across various cancer types for these cohorts remain limited. Here, we expand upon our previous work to generate high-quality protein expression data for approximately 8,000 TCGA patient samples and around 900 CCLE cell line samples, covering 447 clinically relevant proteins, using reverse-phase protein arrays. These protein expression profiles offer profound insights into intertumor heterogeneity and cancer dependency and serve as sensitive functional readouts for somatic alterations. We develop a systematic protein-centered strategy for identifying synthetic lethality pairs and experimentally validate an interaction between protein kinase A subunit α and epidermal growth factor receptor. We also identify metastasis-related protein markers with clinical relevance. This dataset represents a valuable resource for advancing our understanding of cancer mechanisms, discovering protein biomarkers and developing innovative therapeutic strategies. Liang and colleagues establish a high-quality protein expression resource for 8,000 The Cancer Genome Atlas patient samples and 900 Cancer Cell Line Encyclopedia cell lines for approximately 450 proteins, which they use to identify synthetic lethality pairs and metastasis markers.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature cancer
Medicine-Oncology
CiteScore
31.10
自引率
1.80%
发文量
129
期刊介绍:
Cancer is a devastating disease responsible for millions of deaths worldwide. However, many of these deaths could be prevented with improved prevention and treatment strategies. To achieve this, it is crucial to focus on accurate diagnosis, effective treatment methods, and understanding the socioeconomic factors that influence cancer rates.
Nature Cancer aims to serve as a unique platform for sharing the latest advancements in cancer research across various scientific fields, encompassing life sciences, physical sciences, applied sciences, and social sciences. The journal is particularly interested in fundamental research that enhances our understanding of tumor development and progression, as well as research that translates this knowledge into clinical applications through innovative diagnostic and therapeutic approaches. Additionally, Nature Cancer welcomes clinical studies that inform cancer diagnosis, treatment, and prevention, along with contributions exploring the societal impact of cancer on a global scale.
In addition to publishing original research, Nature Cancer will feature Comments, Reviews, News & Views, Features, and Correspondence that hold significant value for the diverse field of cancer research.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


