体内CRISPR筛选确定了MEN1在调节肿瘤与微环境相互作用中的双重功能
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
摘要
二维细胞培养模型中的功能基因组筛选在确定影响肿瘤微环境的治疗靶点方面存在局限性。通过比较二维培养基中的 CRISPR-Cas9 靶向筛选和来自同一细胞系的异种移植,我们发现 MEN1 是在体外和体内产生不同剔除效应的首要靶点。多种实体癌类型的 MEN1 基因敲除不会影响体外的细胞增殖,但会分别显著促进或抑制免疫缺陷小鼠或免疫功能健全小鼠的肿瘤生长。从机理上讲,MEN1 基因敲除会重新分配 MLL1 染色质的占有率,增加重复基因组区域的 H3K4me3,激活双链 RNA 的表达,并分别增加免疫缺陷小鼠和免疫功能正常小鼠的中性粒细胞和 CD8+ T 细胞浸润。药物抑制 menin-MLL 相互作用会以 CD8+ T 细胞依赖的方式减少肿瘤生长。这些发现揭示了 MEN1 依赖于肿瘤微环境的致癌和抑瘤功能,为在实体瘤中靶向治疗 MEN1 提供了理论依据。本文章由计算机程序翻译,如有差异,请以英文原文为准。
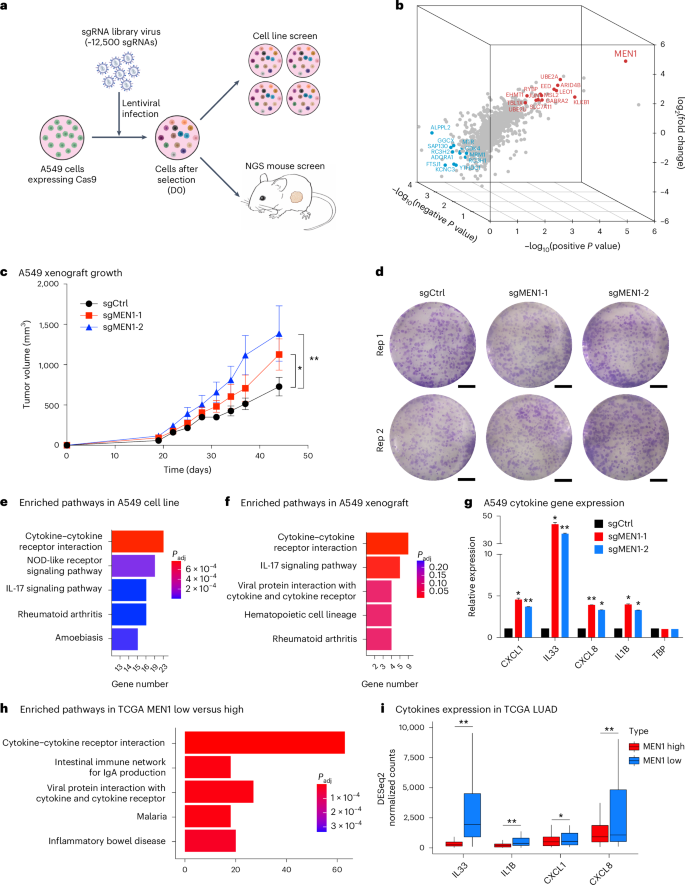
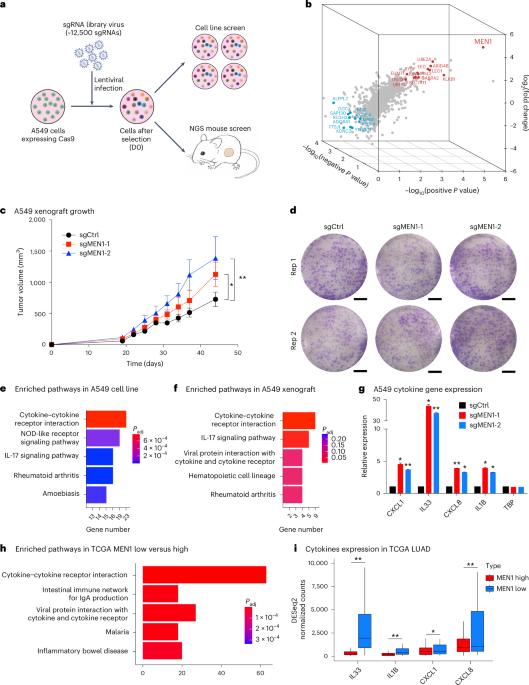
In vivo CRISPR screens identify a dual function of MEN1 in regulating tumor–microenvironment interactions
Functional genomic screens in two-dimensional cell culture models are limited in identifying therapeutic targets that influence the tumor microenvironment. By comparing targeted CRISPR–Cas9 screens in a two-dimensional culture with xenografts derived from the same cell line, we identified MEN1 as the top hit that confers differential dropout effects in vitro and in vivo. MEN1 knockout in multiple solid cancer types does not impact cell proliferation in vitro but significantly promotes or inhibits tumor growth in immunodeficient or immunocompetent mice, respectively. Mechanistically, MEN1 knockout redistributes MLL1 chromatin occupancy, increasing H3K4me3 at repetitive genomic regions, activating double-stranded RNA expression and increasing neutrophil and CD8+ T cell infiltration in immunodeficient and immunocompetent mice, respectively. Pharmacological inhibition of the menin–MLL interaction reduces tumor growth in a CD8+ T cell-dependent manner. These findings reveal tumor microenvironment-dependent oncogenic and tumor-suppressive functions of MEN1 and provide a rationale for targeting MEN1 in solid cancers. Loss of MEN1 affects tumor growth, varing with the components of the tumor microenvironment. These tumors show redistribution of MLL1 on chromatin and the activation of a viral mimicry response.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


