非生产性剪接对人类基因表达的全球影响
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
摘要
人类基因中的替代剪接(AS)被广泛认为是提高蛋白质组多样性的一种机制。通过产生 "非生产性 "转录本,无义介导衰变(NMD)将其作为快速降解的目标,AS 也可以影响基因表达水平而不增加蛋白质多样性。然而,这种调控机制的相对重要性仍未得到充分探索。为了更好地了解 AS-NMD 相对于其他调控机制的影响,我们分析了从转录到细胞质衰变各个阶段的八项分子检测的群体规模基因组数据。与之前使用稳态 RNA 估算的结果相比,我们报告的非生产性剪接增加了三倍。这种非生产性剪接化合物跨越了多基因,导致蛋白质编码基因中 15% 的转录本分子是非生产性的。利用细胞系间的遗传变异,我们发现由 AS 解释的 GWAS 性状相关位点通常与 NMD 诱导的表达水平差异以及蛋白质同工酶的使用差异相关。我们的研究结果表明,AS 的大部分影响是由 NMD 诱导的基因表达变化介导的,而不是蛋白质组的多样化。本文章由计算机程序翻译,如有差异,请以英文原文为准。
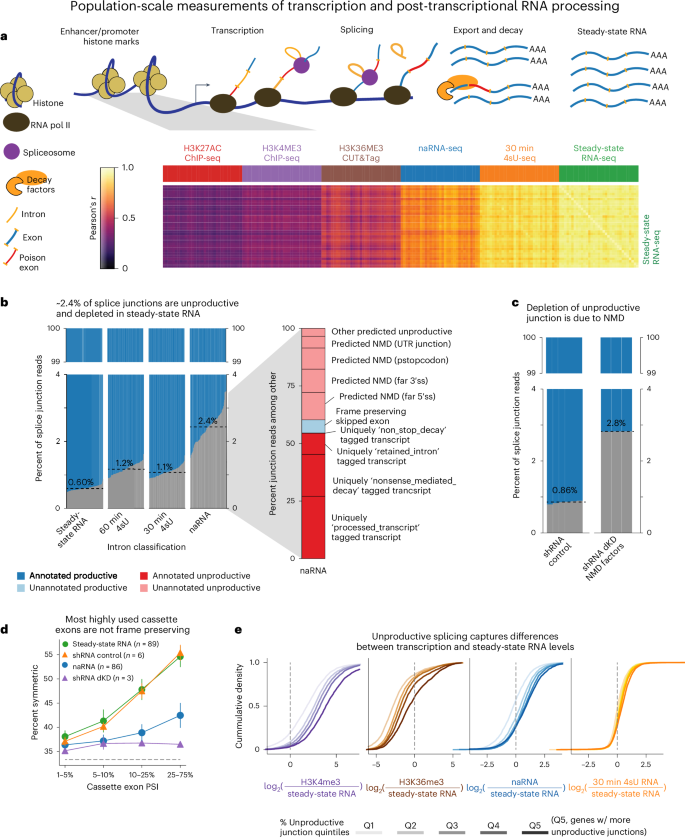
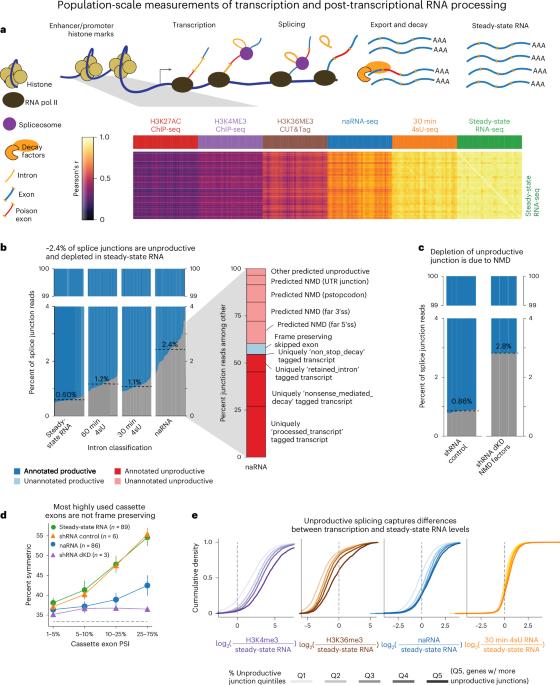
Global impact of unproductive splicing on human gene expression
Alternative splicing (AS) in human genes is widely viewed as a mechanism for enhancing proteomic diversity. AS can also impact gene expression levels without increasing protein diversity by producing ‘unproductive’ transcripts that are targeted for rapid degradation by nonsense-mediated decay (NMD). However, the relative importance of this regulatory mechanism remains underexplored. To better understand the impact of AS–NMD relative to other regulatory mechanisms, we analyzed population-scale genomic data across eight molecular assays, covering various stages from transcription to cytoplasmic decay. We report threefold more unproductive splicing compared with prior estimates using steady-state RNA. This unproductive splicing compounds across multi-intronic genes, resulting in 15% of transcript molecules from protein-coding genes being unproductive. Leveraging genetic variation across cell lines, we find that GWAS trait-associated loci explained by AS are as often associated with NMD-induced expression level differences as with differences in protein isoform usage. Our findings suggest that much of the impact of AS is mediated by NMD-induced changes in gene expression rather than diversification of the proteome. Genomic analyses suggest that ~15% of transcript molecules are spliced into unproductive transcripts targeted by nonsense-mediated decay, which have a larger effect on gene expression than previously thought.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


