机器学习生成的动力学模型能准确描述细胞内的代谢状态
IF 42.8
1区 化学
Q1 CHEMISTRY, PHYSICAL
引用次数: 0
摘要
生成大型全息数据集已成为深入了解细胞过程的例行工作,然而破译这些数据集以确定代谢状态仍具有挑战性。动力学模型可以明确地将代谢物浓度、代谢通量和酶水平联系起来,从而帮助整合全微观数据。然而,确定细胞生理学的动力学参数对这些代谢数学表征的广泛应用构成了明显的障碍。在这里,我们介绍了 RENAISSANCE,这是一种生成式机器学习框架,用于高效地为具有与实验观察相匹配的动态特性的大规模动力学模型设置参数。RENAISSANCE 通过无缝整合各种 omics 数据和其他相关信息(包括细胞外培养基成分、理化数据和领域专家的专业知识),准确描述了大肠杆菌的细胞内代谢状态。它还能估算缺失的动力学参数,并将其与稀少的实验数据相协调,从而大大降低参数的不确定性,提高准确性。这一框架对于研究代谢变化(涉及代谢物和酶水平的变化以及健康和生物技术领域的酶活性)的研究人员非常有价值。本文章由计算机程序翻译,如有差异,请以英文原文为准。
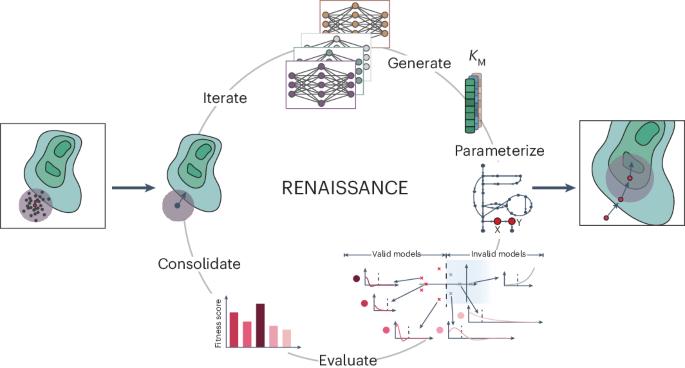
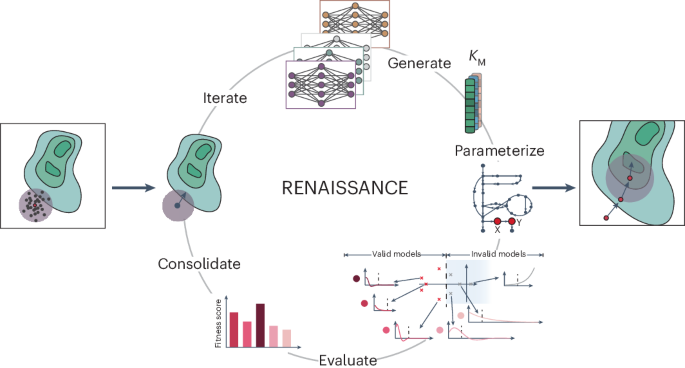
Generative machine learning produces kinetic models that accurately characterize intracellular metabolic states
Generating large omics datasets has become routine for gaining insights into cellular processes, yet deciphering these datasets to determine metabolic states remains challenging. Kinetic models can help integrate omics data by explicitly linking metabolite concentrations, metabolic fluxes and enzyme levels. Nevertheless, determining the kinetic parameters that underlie cellular physiology poses notable obstacles to the widespread use of these mathematical representations of metabolism. Here we present RENAISSANCE, a generative machine learning framework for efficiently parameterizing large-scale kinetic models with dynamic properties matching experimental observations. Through seamless integration of diverse omics data and other relevant information, including extracellular medium composition, physicochemical data and expertise of domain specialists, RENAISSANCE accurately characterizes intracellular metabolic states in Escherichia coli. It also estimates missing kinetic parameters and reconciles them with sparse experimental data, substantially reducing parameter uncertainty and improving accuracy. This framework will be valuable for researchers studying metabolic variations involving changes in metabolite and enzyme levels and enzyme activity in health and biotechnology. Despite the availability of large omics datasets, determining intracellular metabolic states is challenging. Now a generative machine learning framework called RENAISSANCE has been developed to estimate missing kinetic parameters and determine time-resolved metabolic reaction rates and metabolite concentrations without requiring training data.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Nature Catalysis
Chemical Engineering-Bioengineering
CiteScore
52.10
自引率
1.10%
发文量
140
期刊介绍:
Nature Catalysis serves as a platform for researchers across chemistry and related fields, focusing on homogeneous catalysis, heterogeneous catalysis, and biocatalysts, encompassing both fundamental and applied studies. With a particular emphasis on advancing sustainable industries and processes, the journal provides comprehensive coverage of catalysis research, appealing to scientists, engineers, and researchers in academia and industry.
Maintaining the high standards of the Nature brand, Nature Catalysis boasts a dedicated team of professional editors, rigorous peer-review processes, and swift publication times, ensuring editorial independence and quality. The journal publishes work spanning heterogeneous catalysis, homogeneous catalysis, and biocatalysis, covering areas such as catalytic synthesis, mechanisms, characterization, computational studies, nanoparticle catalysis, electrocatalysis, photocatalysis, environmental catalysis, asymmetric catalysis, and various forms of organocatalysis.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


