用合成转录抑制因子直接抑制肿瘤缺氧反应
IF 13.7
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
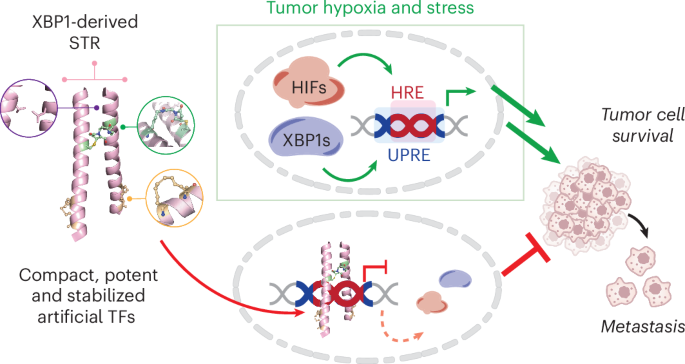
许多致癌转录因子(TFs)被认为是不可药用的,因为它们依赖于大的蛋白质-蛋白质和蛋白质-DNA界面。低氧诱导因子(HIFs)和X-box结合蛋白1(XBP1)等TFs在实体瘤中由低氧和其他压力诱导,并与未折叠蛋白反应元件(UPRE)和低氧诱导反应元件(HRE)基序结合,控制致癌基因程序。在这里,我们报告了一种创建合成转录抑制因子(STR)的策略,这种抑制因子可模仿 XBP1 的基本亮氨酸拉链结构域并识别 UPRE 和 HRE 基序。先导分子 STR22 能高保真地结合 UPRE 和 HRE DNA 序列,并在细胞中与这两种 TF 竞争。在缺氧条件下,STR22 可全面抑制 HIF1α 与含 HRE 启动子和增强子的结合,抑制缺氧诱导的基因表达,并阻断三阴性乳腺癌(TNBC)细胞的原瘤表型。在体内,瘤内和全身 STR22 治疗抑制了 TNBC 肿瘤的低氧依赖基因表达、原发性肿瘤生长和转移。这些数据验证了一种通过协调抑制 TF-DNA 结合来靶向肿瘤缺氧反应的新策略。本文章由计算机程序翻译,如有差异,请以英文原文为准。

Direct inhibition of tumor hypoxia response with synthetic transcriptional repressors
Many oncogenic transcription factors (TFs) are considered to be undruggable because of their reliance on large protein–protein and protein–DNA interfaces. TFs such as hypoxia-inducible factors (HIFs) and X-box-binding protein 1 (XBP1) are induced by hypoxia and other stressors in solid tumors and bind to unfolded protein response element (UPRE) and hypoxia-induced response element (HRE) motifs to control oncogenic gene programs. Here, we report a strategy to create synthetic transcriptional repressors (STRs) that mimic the basic leucine zipper domain of XBP1 and recognize UPRE and HRE motifs. A lead molecule, STR22, binds UPRE and HRE DNA sequences with high fidelity and competes with both TFs in cells. Under hypoxia, STR22 globally suppresses HIF1α binding to HRE-containing promoters and enhancers, inhibits hypoxia-induced gene expression and blocks protumorigenic phenotypes in triple-negative breast cancer (TNBC) cells. In vivo, intratumoral and systemic STR22 treatment inhibited hypoxia-dependent gene expression, primary tumor growth and metastasis of TNBC tumors. These data validate a novel strategy to target the tumor hypoxia response through coordinated inhibition of TF–DNA binding. Qiao and Nguyen et al. describe a strategy to block hypoxia-dependent gene expression in cell and mouse models of breast cancer through dual targeting of X-box-binding protein 1 and hypoxia-inducible factor binding to DNA with fully synthetic, stabilized artificial transcription factors.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature chemical biology
生物-生化与分子生物学
CiteScore
23.90
自引率
1.40%
发文量
238
审稿时长
12 months
期刊介绍:
Nature Chemical Biology stands as an esteemed international monthly journal, offering a prominent platform for the chemical biology community to showcase top-tier original research and commentary. Operating at the crossroads of chemistry, biology, and related disciplines, chemical biology utilizes scientific ideas and approaches to comprehend and manipulate biological systems with molecular precision.
The journal embraces contributions from the growing community of chemical biologists, encompassing insights from chemists applying principles and tools to biological inquiries and biologists striving to comprehend and control molecular-level biological processes. We prioritize studies unveiling significant conceptual or practical advancements in areas where chemistry and biology intersect, emphasizing basic research, especially those reporting novel chemical or biological tools and offering profound molecular-level insights into underlying biological mechanisms.
Nature Chemical Biology also welcomes manuscripts describing applied molecular studies at the chemistry-biology interface due to the broad utility of chemical biology approaches in manipulating or engineering biological systems. Irrespective of scientific focus, we actively seek submissions that creatively blend chemistry and biology, particularly those providing substantial conceptual or methodological breakthroughs with the potential to open innovative research avenues. The journal maintains a robust and impartial review process, emphasizing thorough chemical and biological characterization.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


