细胞外 Ca2+ 激活的核酸酶(EcnA)介导自然能力古菌的转化
IF 2.6
2区 生物学
Q3 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
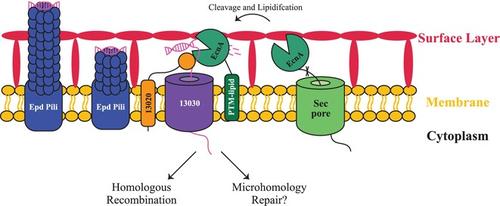
转化(直接从环境中吸收 DNA)是微生物种群基因流动的主要驱动力。在细菌中,DNA 的吸收需要核酸酶将 dsDNA 处理成 ssDNA,然后将其转移到细胞中并纳入基因组。然而,古细菌的 DNA 吸收过程尚不清楚。此前,我们对海洋甲烷球菌(Methanococcus maripaludis)自然转化所必需的基因进行了编目,但几乎没有发现细菌转化相关基因的同源物。在这里,我们确定了一个基因 MMJJ_16440(在此命名为 ecnA)的特征,它是一种细胞外核酸酶。我们发现 EcnA 由 Ca2+ 激活,存在于细胞表面,并且对转化至关重要。虽然 EcnA 能降解多种形式的 DNA,但以 ssDNA 为底物时的活性最高。对环状dsDNA也观察到了活性,这表明EcnA是一种内切酶。这是首次对古菌领域的转化相关蛋白进行生化鉴定,表明古菌和细菌的转化都是以类似的方式开始的。本文章由计算机程序翻译,如有差异,请以英文原文为准。
An Extracellular, Ca2+‐Activated Nuclease (EcnA) Mediates Transformation in a Naturally Competent Archaeon
Transformation, the uptake of DNA directly from the environment, is a major driver of gene flow in microbial populations. In bacteria, DNA uptake requires a nuclease that processes dsDNA to ssDNA, which is subsequently transferred into the cell and incorporated into the genome. However, the process of DNA uptake in archaea is still unknown. Previously, we cataloged genes essential to natural transformation in Methanococcus maripaludis , but few homologs of bacterial transformation‐associated genes were identified. Here, we characterize one gene, MMJJ_16440 (named here as ecnA ), to be an extracellular nuclease. We show that EcnA is Ca2+ ‐activated, present on the cell surface, and essential for transformation. While EcnA can degrade several forms of DNA, the highest activity was observed with ssDNA as a substrate. Activity was also observed with circular dsDNA, suggesting that EcnA is an endonuclease. This is the first biochemical characterization of a transformation‐associated protein in a member of the archaeal domain and suggests that both archaeal and bacterial transformation initiate in an analogous fashion.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Molecular Microbiology
生物-生化与分子生物学
CiteScore
7.20
自引率
5.60%
发文量
132
审稿时长
1.7 months
期刊介绍:
Molecular Microbiology, the leading primary journal in the microbial sciences, publishes molecular studies of Bacteria, Archaea, eukaryotic microorganisms, and their viruses.
Research papers should lead to a deeper understanding of the molecular principles underlying basic physiological processes or mechanisms. Appropriate topics include gene expression and regulation, pathogenicity and virulence, physiology and metabolism, synthesis of macromolecules (proteins, nucleic acids, lipids, polysaccharides, etc), cell biology and subcellular organization, membrane biogenesis and function, traffic and transport, cell-cell communication and signalling pathways, evolution and gene transfer. Articles focused on host responses (cellular or immunological) to pathogens or on microbial ecology should be directed to our sister journals Cellular Microbiology and Environmental Microbiology, respectively.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


