植物表观遗传基因调控及其在作物改良中的潜在应用。
IF 90.2
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
摘要
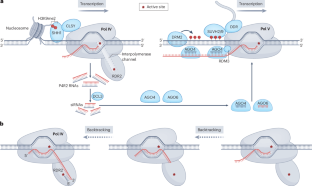
DNA 甲基化又称 5-甲基胞嘧啶,是一种表观遗传修饰,在植物生长、发育和适应过程中具有重要功能。细胞 DNA 甲基化水平受 DNA 甲基转移酶和去甲基化酶的联合作用严格调控。参与 DNA 甲基化靶向和解释的蛋白质复合物已经确定,揭示了甲基-DNA 结合蛋白和分子伴侣的有趣作用。结构研究和体外重组酶系统为 RNA 引导的 DNA 甲基化(植物中催化从头甲基化的主要途径)提供了机制上的见解。更好地了解调控机制将有助于对 DNA 甲基化状态进行特定位点操作。目前正在为此开发基于 CRISPR-dCas9 的表观基因组编辑工具。鉴于 DNA 甲基化模式可通过减数分裂稳定传递,而且表型变异可产生较大的表型变异,表观基因组编辑可在相同的遗传物质上产生额外的表型变异,因而在作物育种方面大有可为。本文章由计算机程序翻译,如有差异,请以英文原文为准。

Epigenetic gene regulation in plants and its potential applications in crop improvement
DNA methylation, also known as 5-methylcytosine, is an epigenetic modification that has crucial functions in plant growth, development and adaptation. The cellular DNA methylation level is tightly regulated by the combined action of DNA methyltransferases and demethylases. Protein complexes involved in the targeting and interpretation of DNA methylation have been identified, revealing intriguing roles of methyl-DNA binding proteins and molecular chaperones. Structural studies and in vitro reconstituted enzymatic systems have provided mechanistic insights into RNA-directed DNA methylation, the main pathway catalysing de novo methylation in plants. A better understanding of the regulatory mechanisms will enable locus-specific manipulation of the DNA methylation status. CRISPR–dCas9-based epigenome editing tools are being developed for this goal. Given that DNA methylation patterns can be stably transmitted through meiosis, and that large phenotypic variations can be contributed by epimutations, epigenome editing holds great promise in crop breeding by creating additional phenotypic variability on the same genetic material. This Review outlines progress in understanding the mechanisms of DNA methylation regulation in plants. Studies in various plants have revealed novel and diverse biological functions of DNA methylation that might assist in developing epigenome editing approaches suitable for crop breeding.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
173.60
自引率
0.50%
发文量
118
审稿时长
6-12 weeks
期刊介绍:
Nature Reviews Molecular Cell Biology is a prestigious journal that aims to be the primary source of reviews and commentaries for the scientific communities it serves. The journal strives to publish articles that are authoritative, accessible, and enriched with easily understandable figures, tables, and other display items. The goal is to provide an unparalleled service to authors, referees, and readers, and the journal works diligently to maximize the usefulness and impact of each article. Nature Reviews Molecular Cell Biology publishes a variety of article types, including Reviews, Perspectives, Comments, and Research Highlights, all of which are relevant to molecular and cell biologists. The journal's broad scope ensures that the articles it publishes reach the widest possible audience.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


