将 T 细胞受体与分化状态分析相结合的质谱法。
IF 27.7
1区 医学
Q1 IMMUNOLOGY
引用次数: 0
摘要
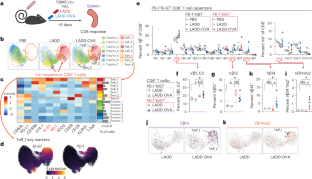
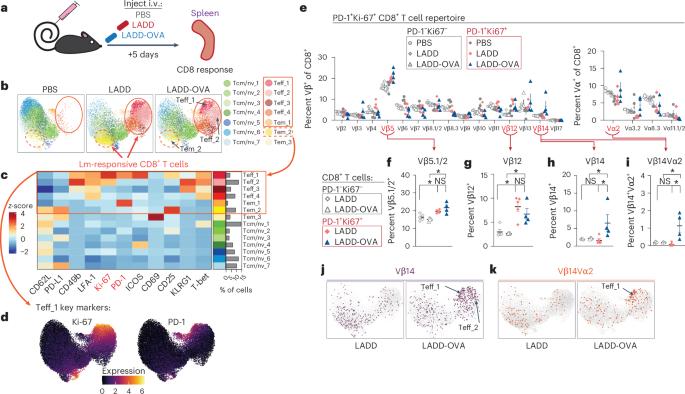
T细胞抗原受体(TCR)识别后的克隆扩增是适应性免疫反应的一个基本特征。在这里,我们提出了一种质量细胞计数(CyTOF)方法,通过将特异性 TCR Vα 和 Vβ 链抗体与小鼠体内 T 细胞活化和分化蛋白抗体相结合来跟踪 T 细胞反应。这种策略能鉴定出表达特定 Vβ 和 Vα 链的 CD8+ 和 CD4+ T 细胞的扩增,它们对李斯特菌、肿瘤和呼吸道流感感染的反应具有不同的分化状态。表达 Vβ 链的 T 细胞扩增可能与识别李斯特菌、肿瘤细胞或流感的特异性抗原直接相关。在流感感染的情况下,我们发现肌肉注射疫苗或康复血清转移等常见治疗方法会改变应答 T 细胞的 TCR 多样性和分化状态。因此,我们提出了一种在适应性免疫反应过程中监测 TCR 使用的广泛变化并同时进行 T 细胞表型分析的方法。本文章由计算机程序翻译,如有差异,请以英文原文为准。
A mass cytometry method pairing T cell receptor and differentiation state analysis
T cell antigen receptor (TCR) recognition followed by clonal expansion is a fundamental feature of adaptive immune responses. Here, we present a mass cytometric (CyTOF) approach to track T cell responses by combining antibodies for specific TCR Vα and Vβ chains with antibodies against T cell activation and differentiation proteins in mice. This strategy identifies expansions of CD8+ and CD4+ T cells expressing specific Vβ and Vα chains with varying differentiation states in response to Listeria monocytogenes, tumors and respiratory influenza infection. Expanded T cell populations expressing Vβ chains could be directly linked to the recognition of specific antigens from Listeria, tumor cells or influenza. In the setting of influenza infection, we found that common therapeutic approaches of intramuscular vaccination or convalescent serum transfer altered the TCR diversity and differentiation state of responding T cells. Thus, we present a method to monitor broad changes in TCR use paired with T cell phenotyping during adaptive immune responses. Here the authors present a mass cytometry-based method for identification of antigen-specific T cells and their differentiation state, testing it in cancer, bacterial and viral mouse models.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Nature Immunology
医学-免疫学
CiteScore
40.00
自引率
2.30%
发文量
248
审稿时长
4-8 weeks
期刊介绍:
Nature Immunology is a monthly journal that publishes the highest quality research in all areas of immunology. The editorial decisions are made by a team of full-time professional editors. The journal prioritizes work that provides translational and/or fundamental insight into the workings of the immune system. It covers a wide range of topics including innate immunity and inflammation, development, immune receptors, signaling and apoptosis, antigen presentation, gene regulation and recombination, cellular and systemic immunity, vaccines, immune tolerance, autoimmunity, tumor immunology, and microbial immunopathology. In addition to publishing significant original research, Nature Immunology also includes comments, News and Views, research highlights, matters arising from readers, and reviews of the literature. The journal serves as a major conduit of top-quality information for the immunology community.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


