绘制空间组织和遗传细胞状态调节因子图,以卵巢癌中的免疫逃避为目标
IF 27.7
1区 医学
Q1 IMMUNOLOGY
引用次数: 0
摘要
免疫逃避的驱动因素尚不完全清楚,这限制了癌症免疫疗法的成功。在这里,我们应用单细胞空间和扰动转录组学来描述高级别浆液性输卵管卵巢癌的免疫逃避。为此,我们首先对来自 94 名患者的 130 个肿瘤中的 250 多万个原位细胞进行了分析,从而绘制了高级别浆液性输卵管卵巢癌的空间组织图。这揭示了反映肿瘤遗传学的恶性细胞状态,并预测了T细胞和自然杀伤细胞的浸润水平以及对免疫检查点阻断的反应。然后,我们进行了 Perturb-seq 筛选,确定了引发这种恶性细胞状态的遗传扰动(包括 PTPN1 和 ACTR8 的敲除)。最后,我们证明这些扰动以及 PTPN1/PTPN2 抑制剂能使卵巢癌细胞对 T 细胞和自然杀伤细胞的细胞毒性敏感,正如预测的那样。因此,这项研究通过将基因变异、细胞状态调节因子和空间生物学联系起来,确定了研究和针对免疫逃避的方法。本文章由计算机程序翻译,如有差异,请以英文原文为准。
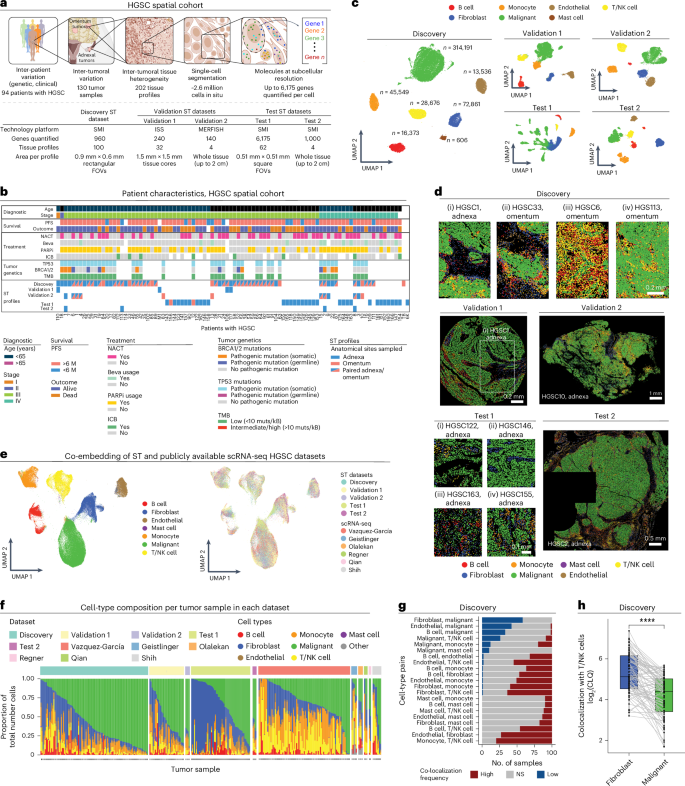
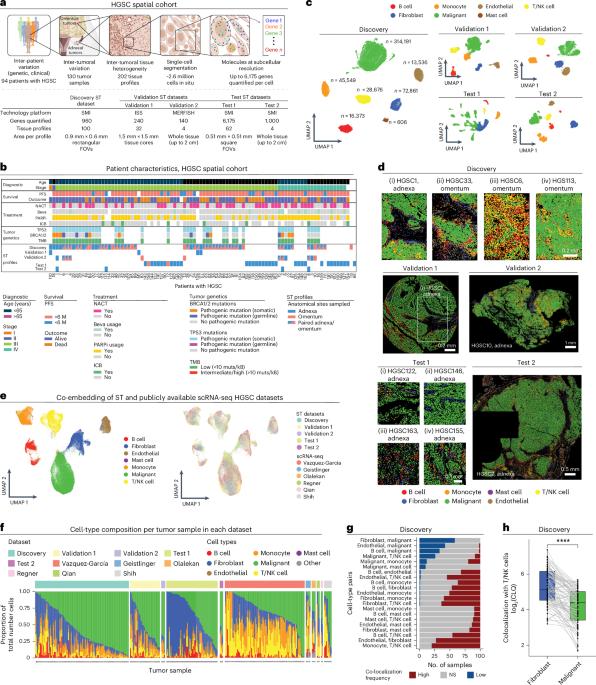
Mapping spatial organization and genetic cell-state regulators to target immune evasion in ovarian cancer
The drivers of immune evasion are not entirely clear, limiting the success of cancer immunotherapies. Here we applied single-cell spatial and perturbational transcriptomics to delineate immune evasion in high-grade serous tubo-ovarian cancer. To this end, we first mapped the spatial organization of high-grade serous tubo-ovarian cancer by profiling more than 2.5 million cells in situ in 130 tumors from 94 patients. This revealed a malignant cell state that reflects tumor genetics and is predictive of T cell and natural killer cell infiltration levels and response to immune checkpoint blockade. We then performed Perturb-seq screens and identified genetic perturbations—including knockout of PTPN1 and ACTR8—that trigger this malignant cell state. Finally, we show that these perturbations, as well as a PTPN1/PTPN2 inhibitor, sensitize ovarian cancer cells to T cell and natural killer cell cytotoxicity, as predicted. This study thus identifies ways to study and target immune evasion by linking genetic variation, cell-state regulators and spatial biology. Here the authors provide a resource for ovarian cancer combining spatial transcriptomics, genomics, CRISPR Perturb-seq screens and in silico methods to focus on T cells and natural killer cells in the tumor and their role in immune evasion.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Nature Immunology
医学-免疫学
CiteScore
40.00
自引率
2.30%
发文量
248
审稿时长
4-8 weeks
期刊介绍:
Nature Immunology is a monthly journal that publishes the highest quality research in all areas of immunology. The editorial decisions are made by a team of full-time professional editors. The journal prioritizes work that provides translational and/or fundamental insight into the workings of the immune system. It covers a wide range of topics including innate immunity and inflammation, development, immune receptors, signaling and apoptosis, antigen presentation, gene regulation and recombination, cellular and systemic immunity, vaccines, immune tolerance, autoimmunity, tumor immunology, and microbial immunopathology. In addition to publishing significant original research, Nature Immunology also includes comments, News and Views, research highlights, matters arising from readers, and reviews of the literature. The journal serves as a major conduit of top-quality information for the immunology community.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


