罕见变异对疾病遗传性的贡献远大于传统估计:等位基因分布模型的修正。
IF 2.6
3区 生物学
Q2 GENETICS & HEREDITY
引用次数: 0
摘要
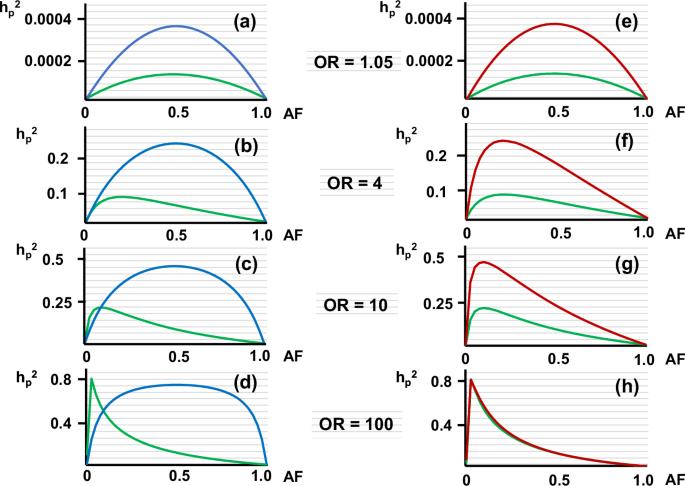
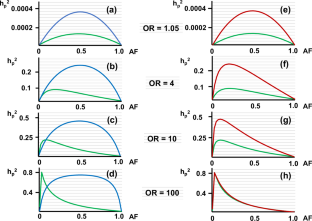
"遗传率缺失 "是人类遗传学目前面临的一个问题。我曾报告过一种方法,无需计算显性和隐性模型下的遗传变异,即可估算常见疾病多态性的遗传率(hp2)。在此,我将该方法扩展到了共显模式,并对 hp2 进行了试算。作为一种常规方法,我还应用 Pawitan 等人最初报告的等位基因分布模型计算 hp2,以进行比较。但出乎意料的是,计算出的高几率罕见变异的 hp2 远远高于等位基因分布模型的计算值。此外,在研究 hp2 计算值差异的基础时,我注意到传统方法使用变异体在普通人群中的等位基因频率(AF)来计算该变异体的遗传变异。然而,这暗含了一个假设,即未受影响的个体包含在疾病的表型中--这一假设与病例对照研究中未受影响的个体属于对照(未受影响)组的假设不一致。因此,我修改了等位基因分布模型,使用了患者群体中的 AF。结果,用修改后的等位基因分布模型计算出的罕见变异的 hp2 相当高。用修改后的等位基因分布模型重新计算文献中报道的几种罕见变异的 hp2,结果是用原始等位基因分布模型计算的 hp2 的 3.2 - 53.7 倍。这些结果表明,罕见变异对疾病遗传性的贡献被大大低估了。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Contribution of rare variants to heritability of a disease is much greater than conventionally estimated: modification of allele distribution model
“Missing heritability” is a current problem in human genetics. I previously reported a method to estimate heritability of a polymorphism (hp2) for a common disease without calculating the genetic variance under dominant and the recessive models. Here, I extend the method to the co-dominant model and carry out trial calculations of hp2. I also calculate hp2 applying the allele distribution model originally reported by Pawitan et al. for comparison as a conventional method. But unexpectedly, hp2 calculated for rare variants with high odds ratios was much higher than the calculated values with the allele distribution model. Also, while examining the basis for the difference in calculated hp2, I noticed that conventional methods use the allele frequency (AF) of a variant in the general population to calculate the genetic variance of that variant. However, this implicitly assumes that the unaffected are included among the phenotypes of the disease – an assumption that is inconsistent with case-control studies in which unaffected individuals belong to the control (unaffected) group. Therefore, I modified the allele distribution model by using the AF in the patient population. Consequently, the hp2 of rare variants calculated with the modified allele distribution model was quite high. Recalculating hp2 of several rare variants reported in the literature with the modified allele distribution model yielded results were 3.2 - 53.7 times higher than the hp2 calculated with the original allele distribution model. These results suggest that the contribution of rare variants to heritability of a disease has been considerably underestimated.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Human Genetics
生物-遗传学
CiteScore
7.20
自引率
0.00%
发文量
101
审稿时长
4-8 weeks
期刊介绍:
The Journal of Human Genetics is an international journal publishing articles on human genetics, including medical genetics and human genome analysis. It covers all aspects of human genetics, including molecular genetics, clinical genetics, behavioral genetics, immunogenetics, pharmacogenomics, population genetics, functional genomics, epigenetics, genetic counseling and gene therapy.
Articles on the following areas are especially welcome: genetic factors of monogenic and complex disorders, genome-wide association studies, genetic epidemiology, cancer genetics, personal genomics, genotype-phenotype relationships and genome diversity.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


