非规范起始密码子赋予小鼠肠道中共生大肠杆菌利用碳水化合物的环境优势
IF 20.5
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
摘要
资源竞争是肠道微生物群组成的一个驱动因素。细菌可以通过限制共用的生长营养物质,在代谢上超越相似的对手。对于具有相同代谢基因集的细菌来说,其背后的机制仍不清楚。在这里,我们分析了小鼠共生大肠杆菌 8178 的乳糖利用操作子。利用体外和体内方法,我们发现乳糖利用抑制基因 lacI 从其原生非规范 GTG 起始密码子开始翻译会增加乳糖利用簇的基础表达,从而增强对乳糖消耗的适应性。因此,携带野生型lacI GTG起始密码子的菌株在小鼠肠道中的表现优于lacI ATG起始密码子突变体。这种优势在通过饮食转移或改变突变体频率来限制宿主乳糖摄入量时会减弱,这强调了单个核苷酸变化对肠道微生物群常见成员的细菌适应性的环境依赖性影响。通过基因组分析,我们的数据揭示了非规范起始密码子在新陈代谢竞争中的一种未被察觉的功能。本文章由计算机程序翻译,如有差异,请以英文原文为准。
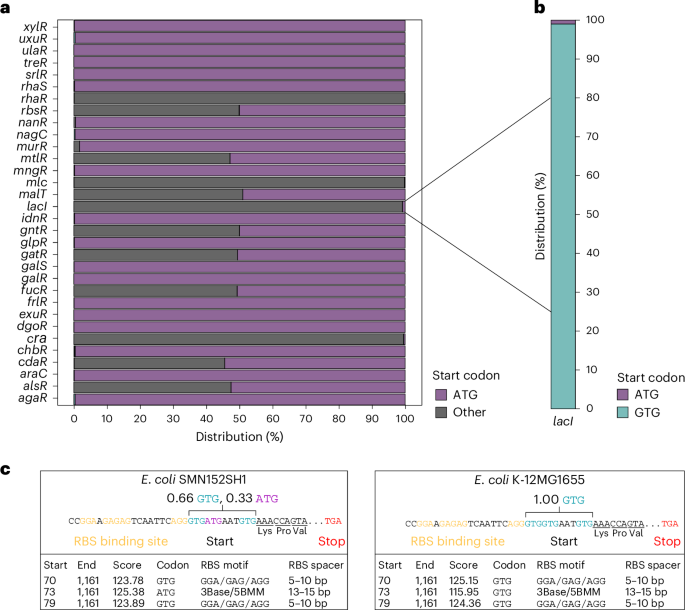
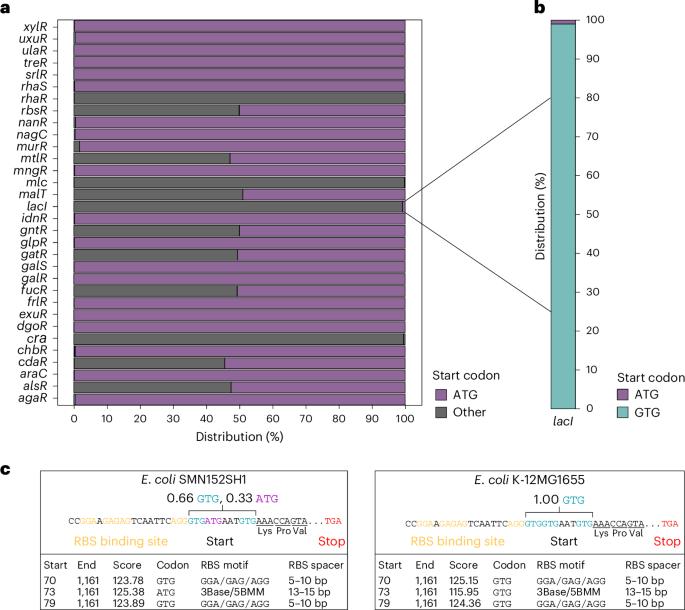
Non-canonical start codons confer context-dependent advantages in carbohydrate utilization for commensal E. coli in the murine gut
Resource competition is a driver of gut microbiota composition. Bacteria can outcompete metabolically similar rivals through the limitation of shared growth-fuelling nutrients. The mechanisms underlying this remain unclear for bacteria with identical sets of metabolic genes. Here we analysed the lactose utilization operon in the murine commensal Escherichia coli 8178. Using in vitro and in vivo approaches, we showed that translation of the lactose utilization repressor gene lacI from its native non-canonical GTG start codon increases the basal expression of the lactose utilization cluster, enhancing adaptation to lactose consumption. Consequently, a strain carrying the wild type lacI GTG start codon outperformed the lacI ATG start codon mutant in the mouse intestine. This advantage was attenuated upon limiting host lactose intake through diet shift or altering the mutant frequency, emphasizing the context-dependent effect of a single nucleotide change on the bacterial fitness of a common member of the gut microbiota. Coupled with a genomic analysis highlighting the selection of non-ATG start codons in sugar utilization regulator genes across the Enterobacteriaceae family, our data exposed an unsuspected function of non-canonical start codons in metabolic competition. Non-canonical start codons promote carbohydrate exploitation and faster metabolic adaptation, conferring growth advantages to commensal Escherichia coli in the mouse gut.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


