染色体级基因组组装揭示了入侵植物葭藻的基因组进化。
IF 5.1
1区 生物学
Q1 BIOLOGY
引用次数: 0
摘要
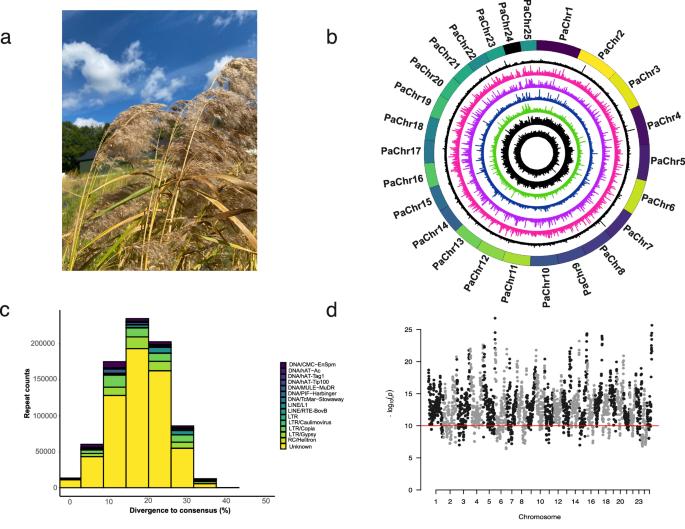
生物入侵对生态系统构成了重大威胁,破坏了当地的生物多样性和生态系统功能。然而,人们对入侵的基因组基础仍然知之甚少,因此很难有效预测和管理入侵物种。普通芦苇(Phragmites australis)是湿地生态系统中的优势草种,从欧洲转移到北美后,其入侵性尤为突出。在这里,我们展示了一个由 24 条假染色体和一条 B 染色体组成的高质量无间隙、端粒到端粒的芦苇基因组。全相位亚基因组显示了相当大的亚基因组优势,并揭示了二倍体祖先在大约 3090 万年前的分化。比较基因组学利用其他三个品系的染色体级支架和以前发表的一个入侵品系的基因组组装草案发现,以串联重复形式出现的基因家族扩张可能是该品系入侵的原因之一。这项研究揭示了 Arundinoideae 禾本科植物的基因组进化,并表明基因家族扩展和串联重复等遗传驱动因素可能是植物生物入侵过程的基础。这些发现为了解和管理植物物种入侵的遗传基础迈出了关键一步。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Chromosome-level genome assemblies reveal genome evolution of an invasive plant Phragmites australis
Biological invasions pose a significant threat to ecosystems, disrupting local biodiversity and ecosystem functions. The genomic underpinnings of invasiveness, however, are still largely unknown, making it difficult to predict and manage invasive species effectively. The common reed (Phragmites australis) is a dominant grass species in wetland ecosystems and has become particularly invasive when transferred from Europe to North America. Here, we present a high-quality gap-free, telomere-to-telomere genome assembly of Phragmites australis consisting of 24 pseudochromosomes and a B chromosome. Fully phased subgenomes demonstrated considerable subgenome dominance and revealed the divergence of diploid progenitors approximately 30.9 million years ago. Comparative genomics using chromosome-level scaffolds for three other lineages and a previously published draft genome assembly of an invasive lineage revealed that gene family expansions in the form of tandem duplications may have contributed to the invasiveness of the lineage. This study sheds light on the genome evolution of Arundinoideae grasses and suggests that genetic drivers, such as gene family expansions and tandem duplications, may underly the processes of biological invasion in plants. These findings provide a crucial step toward understanding and managing the genetic basis of invasiveness in plant species. We provide a complete, telomere to telomere gap-free genome assembly for common reed that includes its B chromosome. Comparative genomic analysis revealed that tandem duplications may have contributed to the invasiveness potential of the species.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Communications Biology
Medicine-Medicine (miscellaneous)
CiteScore
8.60
自引率
1.70%
发文量
1233
审稿时长
13 weeks
期刊介绍:
Communications Biology is an open access journal from Nature Research publishing high-quality research, reviews and commentary in all areas of the biological sciences. Research papers published by the journal represent significant advances bringing new biological insight to a specialized area of research.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


