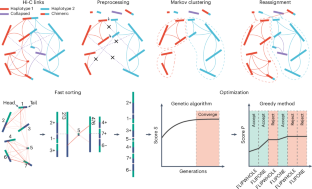
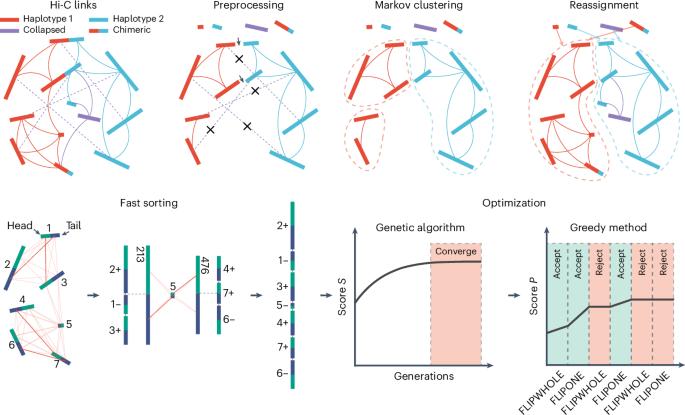
利用 Hi-C 数据实现染色体级单倍型的从头构建。
IF 15.8
1区 生物学
Q1 PLANT SCIENCES
引用次数: 0
摘要
为了克服利用 Hi-C 数据构建染色体级单倍型支架的关键挑战,我们开发了 HapHiC,这是一种 Hi-C 支架工具,在处理单倍型解析的装配时表现出卓越的性能,而无需参考基因组。我们利用 HapHiC 构建了三倍体 Miscanthus × giganteus 的复杂基因组。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Achieving de novo scaffolding of chromosome-level haplotypes using Hi-C data
To overcome key challenges in scaffolding chromosome-level haplotypes with Hi-C data, we developed HapHiC, a Hi-C scaffolding tool that exhibits superior performance in handling haplotype-resolved assemblies without the need for reference genomes. We used HapHiC to construct the complex genome of triploid Miscanthus × giganteus.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Nature Plants
PLANT SCIENCES-
CiteScore
25.30
自引率
2.20%
发文量
196
期刊介绍:
Nature Plants is an online-only, monthly journal publishing the best research on plants — from their evolution, development, metabolism and environmental interactions to their societal significance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


