为纠正异常剪接而设计的定制反义寡核苷酸揭示了罕见遗传疾病的可操作突变群。
IF 9.5
2区 医学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
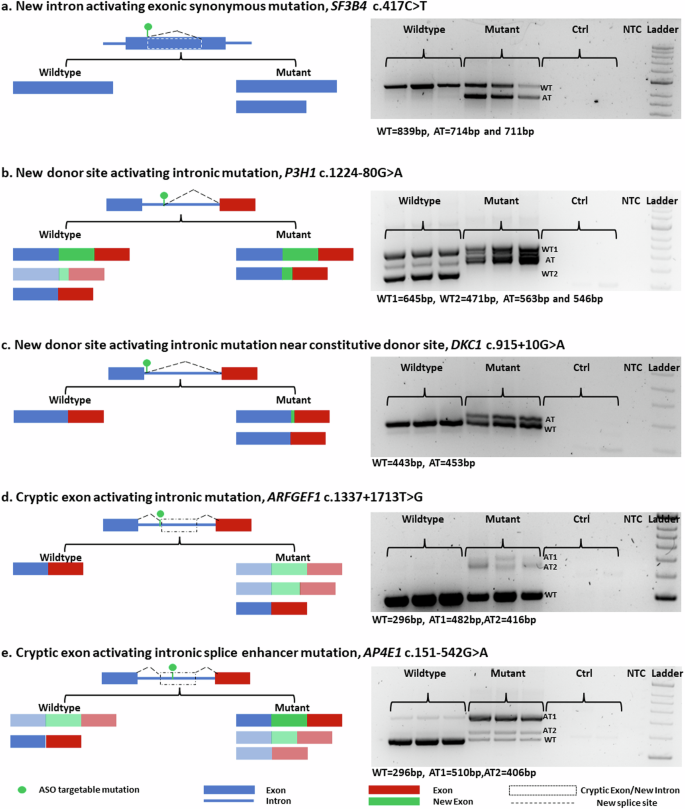
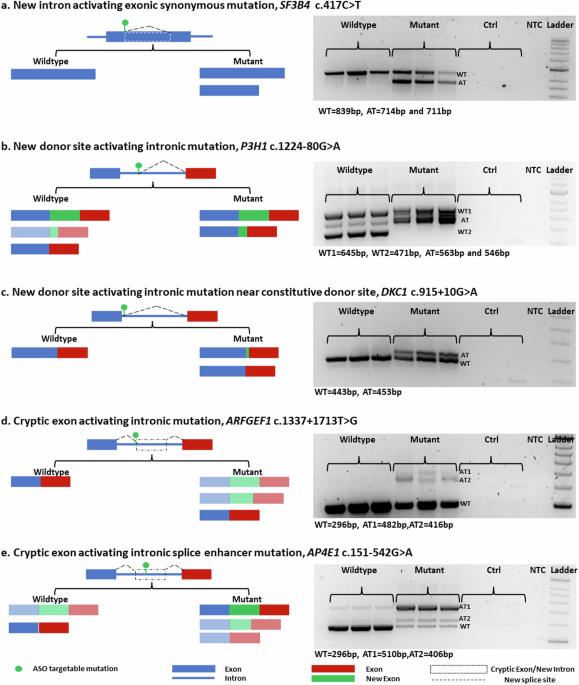
将罕见疾病诊断知识有效地转化为治疗应用可以在合理的时间范围内实现;如果突变适合当前的反义寡核苷酸技术。在我们的研究中,我们在罕见遗传疾病患者中发现了五种不同类型的异常剪接致突变,并针对每种突变类型,使用含或不含八胍树枝状聚合物和 2'-O-methoxyethyl phosphorothioate 的磷酸二铵吗啉寡核苷酸,开发了一种量身定制的反义寡核苷酸。我们观察到处理效果和效率的变化,这既受所选化学成分的影响,也受要校正的异常剪接模式的具体性质的影响。我们的研究表明,所有五种不同类型的异常剪接都能成功纠正。我们的研究结果表明,对异常剪接的有效校正可以依赖于改变寡核苷酸的化学成分,并提出了一种快速、高效、可行的方法,用于在短时间内开发针对遗传疾病的个性化治疗干预措施。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Tailored antisense oligonucleotides designed to correct aberrant splicing reveal actionable groups of mutations for rare genetic disorders
Effective translation of rare disease diagnosis knowledge into therapeutic applications is achievable within a reasonable timeframe; where mutations are amenable to current antisense oligonucleotide technology. In our study, we identified five distinct types of abnormal splice-causing mutations in patients with rare genetic disorders and developed a tailored antisense oligonucleotide for each mutation type using phosphorodiamidate morpholino oligomers with or without octa-guanidine dendrimers and 2′-O-methoxyethyl phosphorothioate. We observed variations in treatment effects and efficiencies, influenced by both the chosen chemistry and the specific nature of the aberrant splicing patterns targeted for correction. Our study demonstrated the successful correction of all five different types of aberrant splicing. Our findings reveal that effective correction of aberrant splicing can depend on altering the chemical composition of oligonucleotides and suggest a fast, efficient, and feasible approach for developing personalized therapeutic interventions for genetic disorders within short time frames. Millions globally suffer from rare diseases, often genetic and affecting children. This study explores using antisense oligonucleotides to fix incorrect RNA splicing, a common result of disease-causing genetic mutations. The results showed that tailored ASOs could correct incorrect splicing for various mutation types, showing this technology′s potential in treating rare genetic diseases. The team chose five mutation types disrupting normal splicing and created specific ASOs to correct these errors in cell models. They created minigenes to simulate the mutations and tested different ASOs′ effectiveness. This method was key to understanding ASOs′ ability to restore normal gene function, crucial for developing targeted treatments for rare genetic disorders. This research could lead to new, targeted treatments for rare genetic disorders, offering hope to millions of patients and their families facing limited treatment options. This summary was initially drafted using artificial intelligence, then revised and fact-checked by the author.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Experimental and Molecular Medicine
医学-生化与分子生物学
CiteScore
19.50
自引率
0.80%
发文量
166
审稿时长
3 months
期刊介绍:
Experimental & Molecular Medicine (EMM) stands as Korea's pioneering biochemistry journal, established in 1964 and rejuvenated in 1996 as an Open Access, fully peer-reviewed international journal. Dedicated to advancing translational research and showcasing recent breakthroughs in the biomedical realm, EMM invites submissions encompassing genetic, molecular, and cellular studies of human physiology and diseases. Emphasizing the correlation between experimental and translational research and enhanced clinical benefits, the journal actively encourages contributions employing specific molecular tools. Welcoming studies that bridge basic discoveries with clinical relevance, alongside articles demonstrating clear in vivo significance and novelty, Experimental & Molecular Medicine proudly serves as an open-access, online-only repository of cutting-edge medical research.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


