使用 clusterProfiler 表征多组学数据。
IF 13.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
随着多组学技术的发展,能够进行多维富集分析的软件对于揭示生物过程和疾病通路中的基因组变化越来越重要。clusterProfiler 因其对数据库的综合利用和先进的可视化功能而脱颖而出。重要的是,clusterProfiler 支持各种生物学知识,包括基因本体论和京都基因和基因组百科全书,通过执行过度呈现和基因组富集分析。一个主要特点是,clusterProfiler 允许用户从各种图形输出中选择可视化结果,从而提高了可解释性。本协议介绍了将 clusterProfiler 用于整合代谢组学和元基因组学分析、鉴定和描述应激条件下的转录因子以及在单细胞研究中注释细胞的创新方法。clusterProfiler 通过 Bioconductor 项目发布,可通过 https://bioconductor.org/packages/clusterProfiler/ 访问。本文章由计算机程序翻译,如有差异,请以英文原文为准。
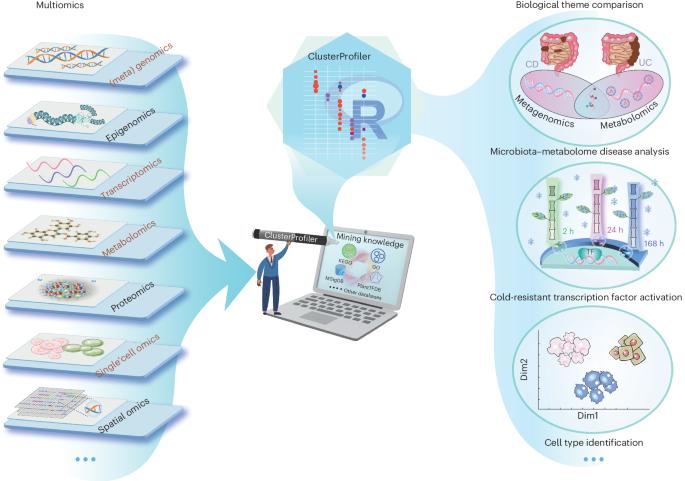
Using clusterProfiler to characterize multiomics data
With the advent of multiomics, software capable of multidimensional enrichment analysis has become increasingly crucial for uncovering gene set variations in biological processes and disease pathways. This is essential for elucidating disease mechanisms and identifying potential therapeutic targets. clusterProfiler stands out for its comprehensive utilization of databases and advanced visualization features. Importantly, clusterProfiler supports various biological knowledge, including Gene Ontology and Kyoto Encyclopedia of Genes and Genomes, through performing over-representation and gene set enrichment analyses. A key feature is that clusterProfiler allows users to choose from various graphical outputs to visualize results, enhancing interpretability. This protocol describes innovative ways in which clusterProfiler has been used for integrating metabolomics and metagenomics analyses, identifying and characterizing transcription factors under stress conditions, and annotating cells in single-cell studies. In all cases, the computational steps can be completed within ~2 min. clusterProfiler is released through the Bioconductor project and can be accessed via https://bioconductor.org/packages/clusterProfiler/ . clusterProfiler is a tool for characterizing and visualizing omics data. The example procedures show integration of metabolomics and metagenomics analyses, characterization of transcription factors and annotation of cells in single-cell studies.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Protocols
生物-生化研究方法
CiteScore
29.10
自引率
0.70%
发文量
128
审稿时长
4 months
期刊介绍:
Nature Protocols focuses on publishing protocols used to address significant biological and biomedical science research questions, including methods grounded in physics and chemistry with practical applications to biological problems. The journal caters to a primary audience of research scientists and, as such, exclusively publishes protocols with research applications. Protocols primarily aimed at influencing patient management and treatment decisions are not featured.
The specific techniques covered encompass a wide range, including but not limited to: Biochemistry, Cell biology, Cell culture, Chemical modification, Computational biology, Developmental biology, Epigenomics, Genetic analysis, Genetic modification, Genomics, Imaging, Immunology, Isolation, purification, and separation, Lipidomics, Metabolomics, Microbiology, Model organisms, Nanotechnology, Neuroscience, Nucleic-acid-based molecular biology, Pharmacology, Plant biology, Protein analysis, Proteomics, Spectroscopy, Structural biology, Synthetic chemistry, Tissue culture, Toxicology, and Virology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


