痛风的发病机制:从基因、表观基因组和转录组研究中获得的分子见解。
IF 29.4
1区 医学
Q1 RHEUMATOLOGY
引用次数: 0
摘要
痛风的发病机制包括一系列步骤,首先是高尿酸血症,其次是尿酸单钠晶体在关节结构中沉积,最后是由 NLRP3 炎性体介导的针对沉积晶体的先天性免疫反应。对血清尿酸水平进行的大型全基因组关联研究(GWAS)初步确定了影响最大的遗传变异,这些变异主要与肾脏和肠道中编码尿酸盐转运体的基因有关。其他全球基因组研究则强调了不常见基因变异的重要性。最近,遗传学和表观遗传学全基因组研究揭示了痛风炎症过程的新途径,包括与表观基因组修饰因子的遗传关联。全表观基因组关联研究还表明,表观基因组重塑与痛风有关联,这或许能调节先天性免疫系统对单钠尿酸盐结晶的反应能力。值得注意的是,痛风全基因组关联研究中涉及的基因并不包括那些编码 NLRP3 炎性体本身成分的基因,而是包括编码参与其调控的分子的基因。通过将基因关联转化为特定的分子机制,人们对痛风的分子机制有了进一步的了解。著名的例子包括 ABCG2、HNF4A、PDZK1、MAF 和 IL37。目前的遗传研究以欧洲血统的参与者为主;然而,针对其他人群的研究正在发现与痛风相关的信息丰富的人群特异性变异。本文章由计算机程序翻译,如有差异,请以英文原文为准。
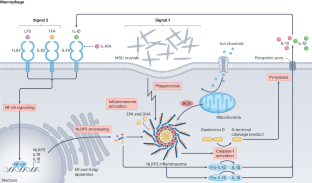
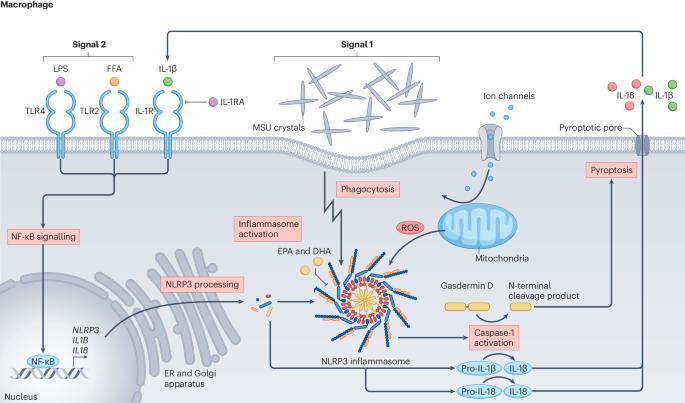
The pathogenesis of gout: molecular insights from genetic, epigenomic and transcriptomic studies
The pathogenesis of gout involves a series of steps beginning with hyperuricaemia, followed by the deposition of monosodium urate crystal in articular structures and culminating in an innate immune response, mediated by the NLRP3 inflammasome, to the deposited crystals. Large genome-wide association studies (GWAS) of serum urate levels initially identified the genetic variants with the strongest effects, mapping mainly to genes that encode urate transporters in the kidney and gut. Other GWAS highlighted the importance of uncommon genetic variants. More recently, genetic and epigenetic genome-wide studies have revealed new pathways in the inflammatory process of gout, including genetic associations with epigenomic modifiers. Epigenome-wide association studies are also implicating epigenomic remodelling in gout, which perhaps regulates the responsiveness of the innate immune system to monosodium urate crystals. Notably, genes implicated in gout GWAS do not include those encoding components of the NLRP3 inflammasome itself, but instead include genes encoding molecules involved in its regulation. Knowledge of the molecular mechanisms underlying gout has advanced through the translation of genetic associations into specific molecular mechanisms. Notable examples include ABCG2, HNF4A, PDZK1, MAF and IL37. Current genetic studies are dominated by participants of European ancestry; however, studies focusing on other population groups are discovering informative population-specific variants associated with gout. Genetic, epigenetic and transcriptomic studies in hyperuricaemia and gout have, in the past 6 years, provided important insights into the underlying molecular mechanisms, revealing new inflammatory pathways and epigenetic factors and expanding research beyond European populations.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Reviews Rheumatology
医学-风湿病学
CiteScore
29.90
自引率
0.90%
发文量
137
审稿时长
6-12 weeks
期刊介绍:
Nature Reviews Rheumatology is part of the Nature Reviews portfolio of journals. The journal scope covers the entire spectrum of rheumatology research. We ensure that our articles are accessible to the widest possible audience.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


