利用 Celloc 准确灵活地绘制从单细胞到空间转录组的图谱
IF 11.1
Q1 MATERIALS SCIENCE, MULTIDISCIPLINARY
引用次数: 0
摘要
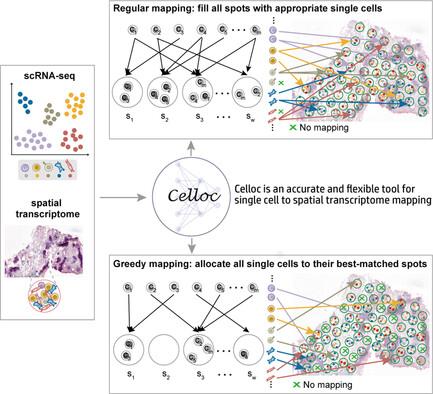
单细胞 RNA 测序(scRNA-seq)与低分辨率空间转录组学(ST)数据之间的精确映射可以弥补 ST 数据的有限分辨率和 scRNA-seq 的空间信息缺失。Celloc 是为此目的开发的一种方法,它结合了图注意自动编码器和综合损失函数,以促进灵活的单细胞到点映射。这样就能在 scRNA-seq 数据中剖析每个点内的细胞组成或分配每个细胞的空间位置。Celloc 的性能以模拟 ST 数据为基准,与最先进的方法相比,具有更高的准确性和鲁棒性。在真实数据集上进行的评估表明,Celloc 可以重建不同组织和组织学区域中各种细胞类型的细胞空间结构。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Accurate and Flexible Single Cell to Spatial Transcriptome Mapping with Celloc
Accurate mapping between single-cell RNA sequencing (scRNA-seq) and low-resolution spatial transcriptomics (ST) data compensates for both limited resolution of ST data and missing spatial information of scRNA-seq. Celloc, a method developed for this purpose, incorporates a graph attention autoencoder and comprehensive loss functions to facilitate flexible single cell-to-spot mapping. This enables either the dissection of cell composition within each spot or the assignment of spatial locations for every cell in scRNA-seq data. Celloc's performance is benchmarked on simulated ST data, demonstrating superior accuracy and robustness compared to state-of-the-art methods. Evaluations on real datasets suggest that Celloc can reconstruct cellular spatial structures with various cell types across different tissues and histological regions.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
14.00
自引率
2.40%
发文量
0
期刊介绍:
Small Science is a premium multidisciplinary open access journal dedicated to publishing impactful research from all areas of nanoscience and nanotechnology. It features interdisciplinary original research and focused review articles on relevant topics. The journal covers design, characterization, mechanism, technology, and application of micro-/nanoscale structures and systems in various fields including physics, chemistry, materials science, engineering, environmental science, life science, biology, and medicine. It welcomes innovative interdisciplinary research and its readership includes professionals from academia and industry in fields such as chemistry, physics, materials science, biology, engineering, and environmental and analytical science. Small Science is indexed and abstracted in CAS, DOAJ, Clarivate Analytics, ProQuest Central, Publicly Available Content Database, Science Database, SCOPUS, and Web of Science.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


