通过归纳转录组学从组织病理学图像预测癌症治疗反应的深度学习框架。
IF 23.5
1区 医学
Q1 ONCOLOGY
引用次数: 0
摘要
人工智能的进步为利用苏木精和伊红染色的肿瘤切片进行精准肿瘤学研究铺平了道路。我们介绍了ENLIGHT-DeepPT,这是一种间接的两步法,包括:(1)DeepPT,这是一种深度学习框架,可预测切片中全基因组肿瘤mRNA的表达;(2)ENLIGHT,可根据推断的表达值预测对靶向疗法和免疫疗法的反应。我们的研究表明,DeepPT 成功预测了所有 16 个已测试的癌症基因组图谱队列的转录组学,并在两个独立的数据集上进行了很好的推广。ENLIGHT-DeepPT成功预测了5个独立患者队列中的真正应答者,涉及6种癌症类型的4种不同治疗方法,总体几率比为2.28,预测应答者的应答率与基线应答率相比提高了39.5%。值得注意的是,它的预测准确率无需对治疗数据进行任何训练即可获得,与直接从图像预测反应的准确率不相上下,后者需要对治疗评估队列进行专门训练。本文章由计算机程序翻译,如有差异,请以英文原文为准。
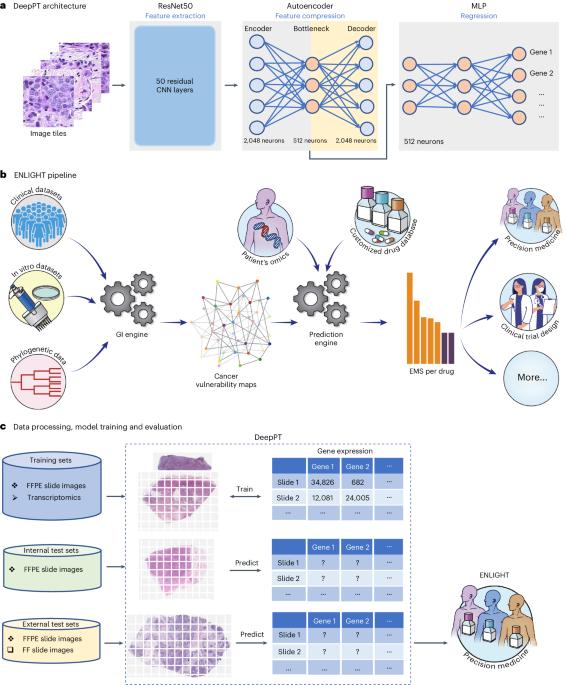
A deep-learning framework to predict cancer treatment response from histopathology images through imputed transcriptomics
Advances in artificial intelligence have paved the way for leveraging hematoxylin and eosin-stained tumor slides for precision oncology. We present ENLIGHT–DeepPT, an indirect two-step approach consisting of (1) DeepPT, a deep-learning framework that predicts genome-wide tumor mRNA expression from slides, and (2) ENLIGHT, which predicts response to targeted and immune therapies from the inferred expression values. We show that DeepPT successfully predicts transcriptomics in all 16 The Cancer Genome Atlas cohorts tested and generalizes well to two independent datasets. ENLIGHT–DeepPT successfully predicts true responders in five independent patient cohorts involving four different treatments spanning six cancer types, with an overall odds ratio of 2.28 and a 39.5% increased response rate among predicted responders versus the baseline rate. Notably, its prediction accuracy, obtained without any training on the treatment data, is comparable to that achieved by directly predicting the response from the images, which requires specific training on the treatment evaluation cohorts. Hoang et al. developed a deep-learning framework called ENLIGHT–DeepPT that predicts therapy response based on imputed transcriptomics and shows predictive power across patient cohorts and cancer types.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature cancer
Medicine-Oncology
CiteScore
31.10
自引率
1.80%
发文量
129
期刊介绍:
Cancer is a devastating disease responsible for millions of deaths worldwide. However, many of these deaths could be prevented with improved prevention and treatment strategies. To achieve this, it is crucial to focus on accurate diagnosis, effective treatment methods, and understanding the socioeconomic factors that influence cancer rates.
Nature Cancer aims to serve as a unique platform for sharing the latest advancements in cancer research across various scientific fields, encompassing life sciences, physical sciences, applied sciences, and social sciences. The journal is particularly interested in fundamental research that enhances our understanding of tumor development and progression, as well as research that translates this knowledge into clinical applications through innovative diagnostic and therapeutic approaches. Additionally, Nature Cancer welcomes clinical studies that inform cancer diagnosis, treatment, and prevention, along with contributions exploring the societal impact of cancer on a global scale.
In addition to publishing original research, Nature Cancer will feature Comments, Reviews, News & Views, Features, and Correspondence that hold significant value for the diverse field of cancer research.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


