cBAF 生成的亚核小体可将 OCT4 的结合范围和功能扩展到增强子的 DNA 主题之外
IF 12.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
典型的 BRG/BRM 相关因子(cBAF)复合物对哺乳动物细胞中增强子的染色质开放至关重要。然而,开放染色质的性质仍不清楚。在这里,我们发现除了产生无组蛋白的 DNA 外,cBAF 还能产生稳定的半球状亚核糖体颗粒,其中包含与 50-80 bp DNA 相关的四个核心组蛋白。我们的全基因组分析表明,cBAF通过靶向和分裂脆弱的核小体来产生这些颗粒。在小鼠胚胎干细胞中,这些亚核小体成为主转录因子OCT4的体内结合底物,与OCT4 DNA基序的存在无关。在增强子上,OCT4与亚核小体的相互作用增加了OCT4的占据率,与无组蛋白DNA占据的区域相比,OCT4结合的基因组间隔扩大了一个数量级。我们认为,依赖于cBAF的亚核小体协调了一种分子机制,将OCT4在染色质开放中的功能投射到其DNA基团之外。本文章由计算机程序翻译,如有差异,请以英文原文为准。
cBAF generates subnucleosomes that expand OCT4 binding and function beyond DNA motifs at enhancers
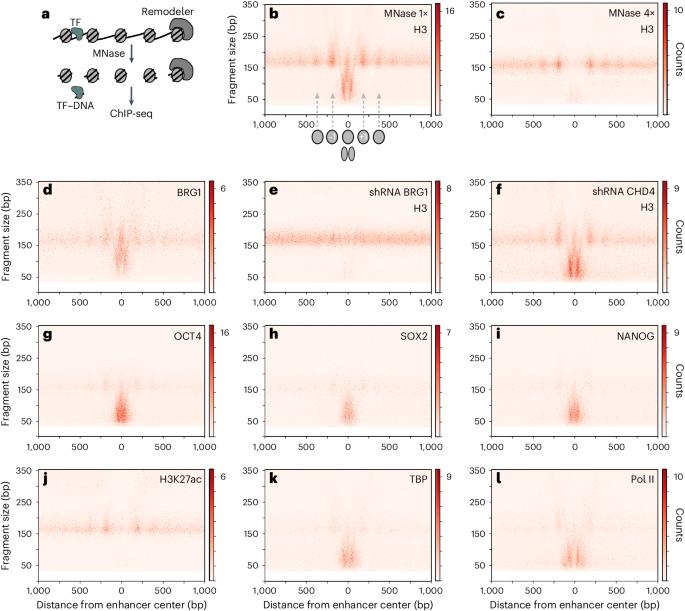
The canonical BRG/BRM-associated factor (cBAF) complex is essential for chromatin opening at enhancers in mammalian cells. However, the nature of the open chromatin remains unclear. Here, we show that, in addition to producing histone-free DNA, cBAF generates stable hemisome-like subnucleosomal particles containing the four core histones associated with 50–80 bp of DNA. Our genome-wide analysis indicates that cBAF makes these particles by targeting and splitting fragile nucleosomes. In mouse embryonic stem cells, these subnucleosomes become an in vivo binding substrate for the master transcription factor OCT4 independently of the presence of OCT4 DNA motifs. At enhancers, the OCT4–subnucleosome interaction increases OCT4 occupancy and amplifies the genomic interval bound by OCT4 by up to one order of magnitude compared to the region occupied on histone-free DNA. We propose that cBAF-dependent subnucleosomes orchestrate a molecular mechanism that projects OCT4 function in chromatin opening beyond its DNA motifs. Here, the authors show that the canonical BRG/BRM-associated factor (SWI/SNF) chromatin remodeler generates subnucleosomes containing 50–80 bp of DNA associated with the four core histones. These hemisome-like particles interact with OCT4 to expand its binding domain at enhancers.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Structural & Molecular Biology
BIOCHEMISTRY & MOLECULAR BIOLOGY-BIOPHYSICS
CiteScore
22.00
自引率
1.80%
发文量
160
审稿时长
3-8 weeks
期刊介绍:
Nature Structural & Molecular Biology is a comprehensive platform that combines structural and molecular research. Our journal focuses on exploring the functional and mechanistic aspects of biological processes, emphasizing how molecular components collaborate to achieve a particular function. While structural data can shed light on these insights, our publication does not require them as a prerequisite.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


