利用运动测序技术确定小鼠行为的结构特征
IF 13.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
小鼠的自发行为是由重复使用的运动模块(如饲养、奔跑或梳理)组成的,这些模块灵活地排列成序列,其内容随时间而变化。通过识别行为模块及其表达顺序,研究人员可以深入了解药物、基因、环境、感官刺激和神经活动对自然行为的影响。我们在此介绍一种执行运动测序(MoSeq)的方案,这是一种受伦理学启发的方法,它利用三维机器视觉和无监督机器学习将小鼠的自发行为分解为一系列称为 "音节 "的元素模块。该协议基于 MoSeq 管道,其中包括深度视频采集、数据预处理和建模模块,以及一套标准化的可视化工具。我们向用户提供了构建 MoSeq 成像设备和获取小鼠自发行为三维视频的说明和代码,以便将其提交给建模框架;该协议的输出包括视频数据每一帧的音节标签,以及描述每个音节使用频率和音节之间过渡情况的汇总图。此外,我们还提供了对关键点-MoSeq 输出进行分析和可视化的说明,MoSeq 是最近开发的一种 MoSeq 变体,可以从标准(而非深度)视频中识别出的关键点中识别出行为主题。这个协议和随附的管道大大降低了门槛,让没有丰富计算伦理学经验的用户也能采用这种无监督、数据驱动的方法来描述小鼠的行为特征。本文章由计算机程序翻译,如有差异,请以英文原文为准。
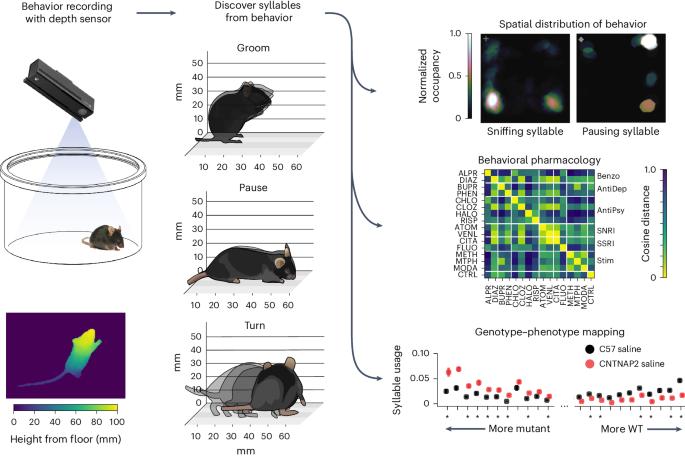
Characterizing the structure of mouse behavior using Motion Sequencing
Spontaneous mouse behavior is composed from repeatedly used modules of movement (e.g., rearing, running or grooming) that are flexibly placed into sequences whose content evolves over time. By identifying behavioral modules and the order in which they are expressed, researchers can gain insight into the effect of drugs, genes, context, sensory stimuli and neural activity on natural behavior. Here we present a protocol for performing Motion Sequencing (MoSeq), an ethologically inspired method that uses three-dimensional machine vision and unsupervised machine learning to decompose spontaneous mouse behavior into a series of elemental modules called ‘syllables’. This protocol is based upon a MoSeq pipeline that includes modules for depth video acquisition, data preprocessing and modeling, as well as a standardized set of visualization tools. Users are provided with instructions and code for building a MoSeq imaging rig and acquiring three-dimensional video of spontaneous mouse behavior for submission to the modeling framework; the outputs of this protocol include syllable labels for each frame of the video data as well as summary plots describing how often each syllable was used and how syllables transitioned from one to the other. In addition, we provide instructions for analyzing and visualizing the outputs of keypoint-MoSeq, a recently developed variant of MoSeq that can identify behavioral motifs from keypoints identified from standard (rather than depth) video. This protocol and the accompanying pipeline significantly lower the bar for users without extensive computational ethology experience to adopt this unsupervised, data-driven approach to characterize mouse behavior. Motion Sequencing uses three-dimensional machine vision and unsupervised machine learning on depth videos to decompose spontaneous mouse behavior into a series of elemental modules called ‘syllables’, revealing how often syllables are used and how they transition over time.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Protocols
生物-生化研究方法
CiteScore
29.10
自引率
0.70%
发文量
128
审稿时长
4 months
期刊介绍:
Nature Protocols focuses on publishing protocols used to address significant biological and biomedical science research questions, including methods grounded in physics and chemistry with practical applications to biological problems. The journal caters to a primary audience of research scientists and, as such, exclusively publishes protocols with research applications. Protocols primarily aimed at influencing patient management and treatment decisions are not featured.
The specific techniques covered encompass a wide range, including but not limited to: Biochemistry, Cell biology, Cell culture, Chemical modification, Computational biology, Developmental biology, Epigenomics, Genetic analysis, Genetic modification, Genomics, Imaging, Immunology, Isolation, purification, and separation, Lipidomics, Metabolomics, Microbiology, Model organisms, Nanotechnology, Neuroscience, Nucleic-acid-based molecular biology, Pharmacology, Plant biology, Protein analysis, Proteomics, Spectroscopy, Structural biology, Synthetic chemistry, Tissue culture, Toxicology, and Virology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


