平行降解组测序和 DMS-MaPseq 大幅修订拟南芥 miRNA 生物发生图谱
IF 13.6
1区 生物学
Q1 PLANT SCIENCES
引用次数: 0
摘要
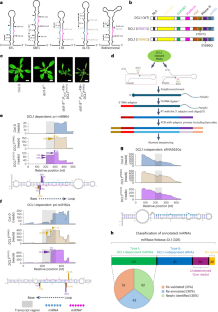
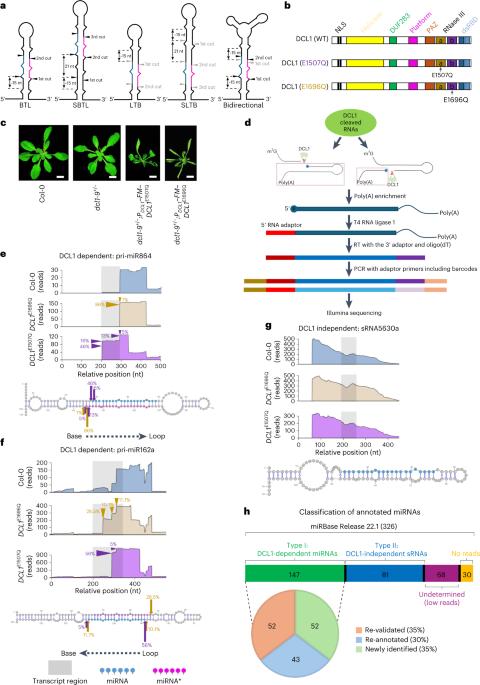
微小核糖核酸(miRNA)由高度结构化的初级转录本(pri-miRNA)产生,调控真核生物中的许多生物过程。由于这些结构的极端异质性,人们已经预测了许多 miRNAs 的植物 pri-miRNAs 的初始加工位点以及决定其加工的结构规则,但对其他 miRNAs 来说仍然难以捉摸。在这里,我们利用半活性 DCL1 突变体和先进的降解测序策略,准确鉴定了之前注释的 326 个拟南芥 miRNA 中 147 个的初始加工位点,并说明了它们相关的 pri-miRNA 裂解模式。对73个pri-miRNA的体内RNA二级结构的阐明表明,其中约95%的pri-miRNA与硅学预测结果不同,修订后的结构对加工位点和模式提供了更清晰的解释。最后,DCL1的伙伴Serrate和HYL1可以协同并独立地影响pri-miRNA的加工模式和体内RNA二级结构。总之,我们的工作揭示了植物 pri-miRNA 的精确加工机制。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Parallel degradome-seq and DMS-MaPseq substantially revise the miRNA biogenesis atlas in Arabidopsis
MicroRNAs (miRNAs) are produced from highly structured primary transcripts (pri-miRNAs) and regulate numerous biological processes in eukaryotes. Due to the extreme heterogeneity of these structures, the initial processing sites of plant pri-miRNAs and the structural rules that determine their processing have been predicted for many miRNAs but remain elusive for others. Here we used semi-active DCL1 mutants and advanced degradome-sequencing strategies to accurately identify the initial processing sites for 147 of 326 previously annotated Arabidopsis miRNAs and to illustrate their associated pri-miRNA cleavage patterns. Elucidating the in vivo RNA secondary structures of 73 pri-miRNAs revealed that about 95% of them differ from in silico predictions, and that the revised structures offer clearer interpretation of the processing sites and patterns. Finally, DCL1 partners Serrate and HYL1 could synergistically and independently impact processing patterns and in vivo RNA secondary structures of pri-miRNAs. Together, our work sheds light on the precise processing mechanisms of plant pri-miRNAs. Parallel degradome sequencing and DMS-MaPseq pinpoint the first cleavage sites on bona fide pri-miRNAs, decode their in vivo structure and provide better interpretation of the cleavage modes and impact of DCL1 cofactors in the process in Arabidopsis.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Nature Plants
PLANT SCIENCES-
CiteScore
25.30
自引率
2.20%
发文量
196
期刊介绍:
Nature Plants is an online-only, monthly journal publishing the best research on plants — from their evolution, development, metabolism and environmental interactions to their societal significance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


