ɑ-螺旋桶的合理种子计算蛋白质设计
IF 12.9
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
计算蛋白质设计进展迅速。在这里,我们描述了从经过验证的平行肽和反平行肽组装开始,设计两个具有结合小分子的中心通道的α螺旋桶状蛋白家族的有效途径。计算设计以已定义的新寡聚桶形肽的序列和结构为种子,通过构建环路将相邻螺旋连接起来。对于具有反平行螺旋的目标,短环就足够了。然而,具有平行螺旋的目标则需要较长的连接,即螺旋-螺旋-螺旋-螺旋图案的外层,这些图案被包装到桶上。在这些计算管道中,定义筒状结构开放状态的残基始终保持不变。这最大限度地减少了序列取样,加快了设计过程。对于六个目标中的每一个目标,只需制造两到六个合成基因在大肠杆菌中表达。平均而言,70% 的合成基因都能表达出可溶性单体蛋白,这些蛋白的特征得到了充分的描述,包括大多数目标物的高分辨率结构,这些结构与设计模型高度吻合。本文章由计算机程序翻译,如有差异,请以英文原文为准。
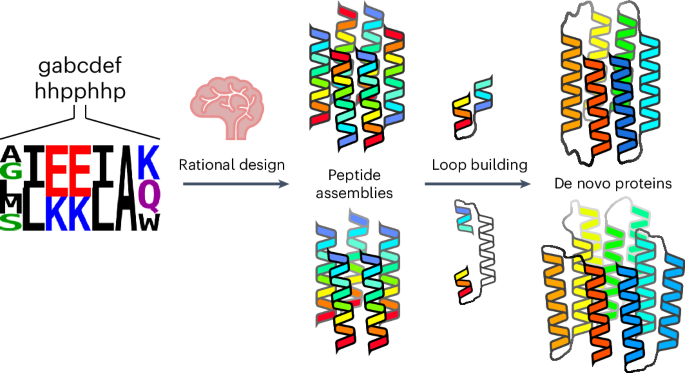
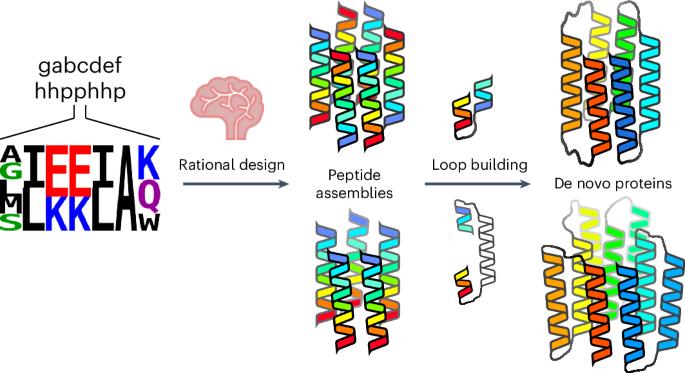
Rationally seeded computational protein design of ɑ-helical barrels
Computational protein design is advancing rapidly. Here we describe efficient routes starting from validated parallel and antiparallel peptide assemblies to design two families of α-helical barrel proteins with central channels that bind small molecules. Computational designs are seeded by the sequences and structures of defined de novo oligomeric barrel-forming peptides, and adjacent helices are connected by loop building. For targets with antiparallel helices, short loops are sufficient. However, targets with parallel helices require longer connectors; namely, an outer layer of helix–turn–helix–turn–helix motifs that are packed onto the barrels. Throughout these computational pipelines, residues that define open states of the barrels are maintained. This minimizes sequence sampling, accelerating the design process. For each of six targets, just two to six synthetic genes are made for expression in Escherichia coli. On average, 70% of these genes express to give soluble monomeric proteins that are fully characterized, including high-resolution structures for most targets that match the design models with high accuracy. An efficient computational pipeline starting from validated peptide assemblies has been used to design two families of α-helical barrel proteins with functionalizable channels. This rationally seeded computational protein design approach delivers soluble, monomeric proteins that match the design targets accurately and with high success rates.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature chemical biology
生物-生化与分子生物学
CiteScore
23.90
自引率
1.40%
发文量
238
审稿时长
12 months
期刊介绍:
Nature Chemical Biology stands as an esteemed international monthly journal, offering a prominent platform for the chemical biology community to showcase top-tier original research and commentary. Operating at the crossroads of chemistry, biology, and related disciplines, chemical biology utilizes scientific ideas and approaches to comprehend and manipulate biological systems with molecular precision.
The journal embraces contributions from the growing community of chemical biologists, encompassing insights from chemists applying principles and tools to biological inquiries and biologists striving to comprehend and control molecular-level biological processes. We prioritize studies unveiling significant conceptual or practical advancements in areas where chemistry and biology intersect, emphasizing basic research, especially those reporting novel chemical or biological tools and offering profound molecular-level insights into underlying biological mechanisms.
Nature Chemical Biology also welcomes manuscripts describing applied molecular studies at the chemistry-biology interface due to the broad utility of chemical biology approaches in manipulating or engineering biological systems. Irrespective of scientific focus, we actively seek submissions that creatively blend chemistry and biology, particularly those providing substantial conceptual or methodological breakthroughs with the potential to open innovative research avenues. The journal maintains a robust and impartial review process, emphasizing thorough chemical and biological characterization.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


