用基因编码的 2,3-二氨基丙酸捕捉酰基酶中间体,用于水解酶底物鉴定。
IF 13.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
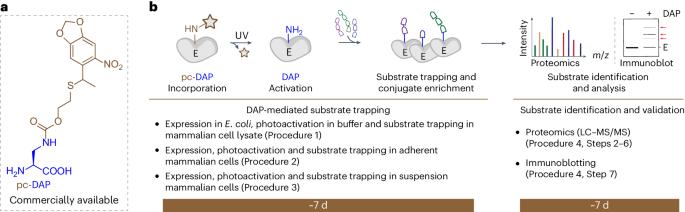
最近开发出了基于催化机理的光激活诱捕器,用于鉴定半胱氨酸或丝氨酸水解酶的底物。这些捕获器是水解酶突变体,其催化半胱氨酸或丝氨酸被基因编码的 2,3-二氨基丙酸(DAP)取代。含 DAP 的水解酶能特异性地捕获蛋白水解反应第一步产生的瞬时硫酯或酯键酰基酶中间产物,使其成为稳定的酰胺类似物。被捕获的底物片段可通过质谱法和免疫印迹法进行水解酶底物的下游鉴定。在本方案中,我们提供了在哺乳动物细胞裂解液和活哺乳动物细胞中捕获底物和鉴定人类疱疹病毒 1 的大护膜蛋白变性肽酶(UL36USP)肽酶结构域的详细步骤指南。其中包括四个步骤:程序 1:哺乳动物细胞裂解物中 DAP 介导的底物捕获(约 8 d);程序 2:粘附哺乳动物细胞中 DAP 介导的底物捕获(约 6 d);程序 3:悬浮哺乳动物细胞中 DAP 介导的底物捕获(约 5 d);程序 4:底物鉴定和验证(约 12-13 d)。实施该方案需要具备在细菌或哺乳动物细胞中进行蛋白质表达、亲和富集和蛋白质组分析的基本技能。本方案将为丝氨酸或半胱氨酸水解酶底物的鉴定提供实用指南,无论是在遗传操作具有挑战性的复杂混合物中,还是在细菌、酵母和哺乳动物细胞等活细胞中。本文章由计算机程序翻译,如有差异,请以英文原文为准。

Capturing acyl–enzyme intermediates with genetically encoded 2,3-diaminopropionic acid for hydrolase substrate identification
Catalytic mechanism-based, light-activated traps have recently been developed to identify the substrates of cysteine or serine hydrolases. These traps are hydrolase mutants whose catalytic cysteine or serine are replaced with genetically encoded 2,3-diaminopropionic acid (DAP). DAP-containing hydrolases specifically capture the transient thioester- or ester-linked acyl–enzyme intermediates resulting from the first step of the proteolytic reaction as their stable amide analogs. The trapped substrate fragments allow the downstream identification of hydrolase substrates by mass spectrometry and immunoblotting. In this protocol, we provide a detailed step-by-step guide for substrate capture and identification of the peptidase domain of the large tegument protein deneddylase (UL36USP) from human herpesvirus 1, both in mammalian cell lysate and live mammalian cells. Four procedures are included: Procedure 1, DAP-mediated substrate trapping in mammalian cell lysate (~8 d); Procedure 2, DAP-mediated substrate trapping in adherent mammalian cells (~6 d); Procedure 3, DAP-mediated substrate trapping in suspension mammalian cells (~5 d); and Procedure 4, substrate identification and validation (~12–13 d). Basic skills to perform protein expression in bacteria or mammalian cells, affinity enrichment and proteomic analysis are required to implement the protocol. This protocol will be a practical guide for identifying substrates of serine or cysteine hydrolases either in a complex mixture, where genetic manipulation is challenging, or in live cells such as bacteria, yeasts and mammalian cells. Light-activated, 2,3-diaminopropionic acid-containing hydrolases trap substrate fragments, facilitating the discovery of new substrates and activities of enzymes in complex mixtures and live cells by mass spectrometry.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Protocols
生物-生化研究方法
CiteScore
29.10
自引率
0.70%
发文量
128
审稿时长
4 months
期刊介绍:
Nature Protocols focuses on publishing protocols used to address significant biological and biomedical science research questions, including methods grounded in physics and chemistry with practical applications to biological problems. The journal caters to a primary audience of research scientists and, as such, exclusively publishes protocols with research applications. Protocols primarily aimed at influencing patient management and treatment decisions are not featured.
The specific techniques covered encompass a wide range, including but not limited to: Biochemistry, Cell biology, Cell culture, Chemical modification, Computational biology, Developmental biology, Epigenomics, Genetic analysis, Genetic modification, Genomics, Imaging, Immunology, Isolation, purification, and separation, Lipidomics, Metabolomics, Microbiology, Model organisms, Nanotechnology, Neuroscience, Nucleic-acid-based molecular biology, Pharmacology, Plant biology, Protein analysis, Proteomics, Spectroscopy, Structural biology, Synthetic chemistry, Tissue culture, Toxicology, and Virology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


