通过分子去灭绝深度学习发现抗生素
IF 26.8
1区 医学
Q1 ENGINEERING, BIOMEDICAL
引用次数: 0
摘要
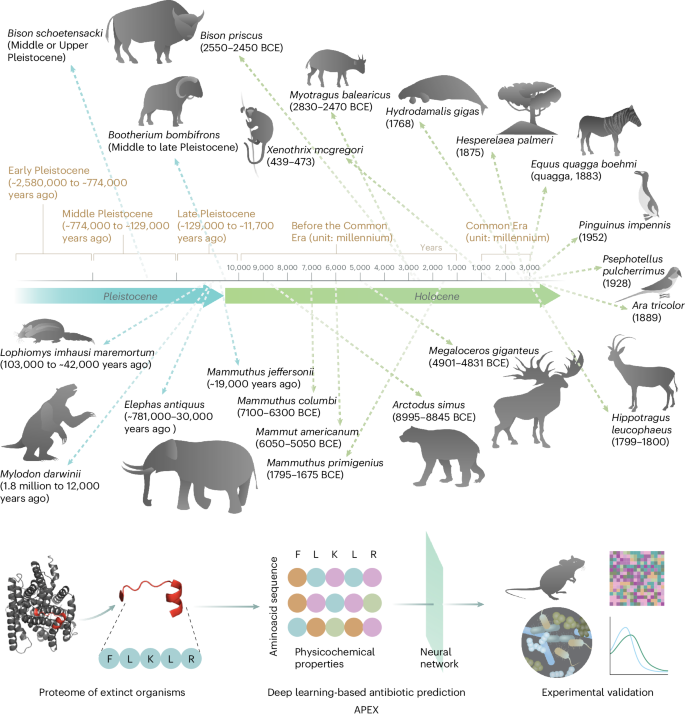
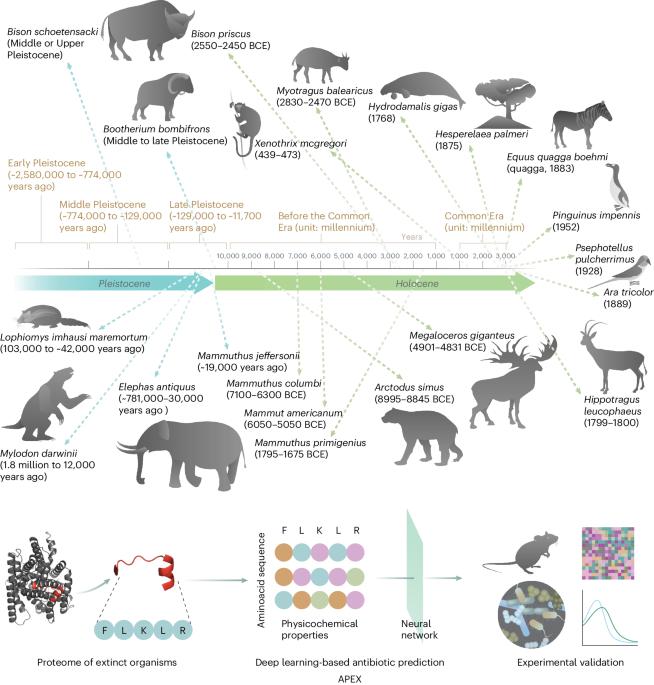
分子灭绝旨在复活分子,以解决抗生素耐药性和其他当今生物与生物医学问题。在这里,我们展示了深度学习可用于挖掘所有已灭绝生物的蛋白质组,以发现抗生素肽。我们训练了由肽序列编码器和神经网络组成的深度学习模型集合,用于预测抗菌活性,并用它挖掘了10,311,899种肽。这些模型预测了 37176 个具有广谱抗菌活性的序列,其中有 11035 个序列在现存生物中没有发现。我们合成了 69 种肽,并通过实验证实了它们对细菌病原体的活性。大多数肽通过使细菌的细胞质膜去极化来杀死细菌,这与已知的抗菌肽相反,后者往往以外膜为目标。值得注意的是,主要化合物(包括长毛猛犸象的mammuthusin-2、直齿象的elephasin-2、古海牛的hydrodamin-1、巨树懒的mylodonin-2和已灭绝的巨麋鹿的megalocerin-1)在皮肤脓肿或大腿感染的小鼠身上显示出抗感染活性。深度学习辅助下的分子去灭绝可能会加速治疗分子的发现。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Deep-learning-enabled antibiotic discovery through molecular de-extinction
Molecular de-extinction aims at resurrecting molecules to solve antibiotic resistance and other present-day biological and biomedical problems. Here we show that deep learning can be used to mine the proteomes of all available extinct organisms for the discovery of antibiotic peptides. We trained ensembles of deep-learning models consisting of a peptide-sequence encoder coupled with neural networks for the prediction of antimicrobial activity and used it to mine 10,311,899 peptides. The models predicted 37,176 sequences with broad-spectrum antimicrobial activity, 11,035 of which were not found in extant organisms. We synthesized 69 peptides and experimentally confirmed their activity against bacterial pathogens. Most peptides killed bacteria by depolarizing their cytoplasmic membrane, contrary to known antimicrobial peptides, which tend to target the outer membrane. Notably, lead compounds (including mammuthusin-2 from the woolly mammoth, elephasin-2 from the straight-tusked elephant, hydrodamin-1 from the ancient sea cow, mylodonin-2 from the giant sloth and megalocerin-1 from the extinct giant elk) showed anti-infective activity in mice with skin abscess or thigh infections. Molecular de-extinction aided by deep learning may accelerate the discovery of therapeutic molecules. Deep learning can be used to mine the proteomes of all available extinct organisms for the discovery of compounds with antibiotic properties.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Biomedical Engineering
Medicine-Medicine (miscellaneous)
CiteScore
45.30
自引率
1.10%
发文量
138
期刊介绍:
Nature Biomedical Engineering is an online-only monthly journal that was launched in January 2017. It aims to publish original research, reviews, and commentary focusing on applied biomedicine and health technology. The journal targets a diverse audience, including life scientists who are involved in developing experimental or computational systems and methods to enhance our understanding of human physiology. It also covers biomedical researchers and engineers who are engaged in designing or optimizing therapies, assays, devices, or procedures for diagnosing or treating diseases. Additionally, clinicians, who make use of research outputs to evaluate patient health or administer therapy in various clinical settings and healthcare contexts, are also part of the target audience.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


