鼻息肉中人类嗜酸性粒细胞的单细胞 RNA 测序揭示了慢性鼻炎组织中嗜酸性粒细胞的异质性
IF 11.4
1区 医学
Q1 ALLERGY
引用次数: 0
摘要
背景在美国,伴有鼻息肉的慢性鼻窦炎(CRSwNP)以2型炎症为特征,但嗜酸性粒细胞在CRSwNP中的实际作用在很大程度上仍不清楚。为了揭示嗜酸性粒细胞在鼻息肉(NP)组织中的作用和异质性,我们对 NP 组织进行了单细胞 RNA 测序(scRNA-Seq)分析。在进行 scRNA-Seq 处理之前,对嗜酸性粒细胞进行了富集。通过微孔 scRNA-Seq 技术(BD Rhapsody 平台)测定了嗜酸性粒细胞的基因表达谱。我们通过基因本体论(geneontology.org)富集和通路分析预测了 NP 嗜酸性粒细胞的整体功能,并通过流式细胞仪确认了所选基因的表达。结果在过滤掉污染细胞后,我们通过 scRNA-Seq 检测到了来自对照 PB 的 5542 个嗜酸性粒细胞、来自 CRSwNP PB 的 3883 个嗜酸性粒细胞、来自对照鼻窦组织的 101 个嗜酸性粒细胞(未纳入进一步分析)以及来自 NPs 的 9727 个嗜酸性粒细胞。我们发现,与所有 PB 嗜酸性粒细胞相比,NP 嗜酸性粒细胞中有 204 个基因下调,354 个基因上调(>1.5 倍,Padj <.05)。NP嗜酸性粒细胞中上调的基因与活化、细胞因子介导的信号转导、生长因子活性、NF-κB信号转导和抗凋亡分子有关。NP嗜酸性粒细胞显示出4个集群,揭示了NP组织中嗜酸性粒细胞的潜在异质性。结论NP组织中升高的嗜酸性粒细胞似乎存在多种亚型,它们可能在CRSwNP中发挥重要的致病作用,部分原因是它们控制了炎症和其他细胞的过度增殖。本文章由计算机程序翻译,如有差异,请以英文原文为准。
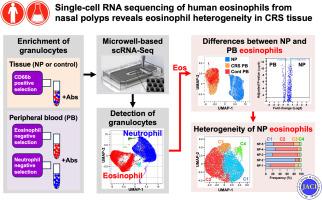
Single cell RNA sequencing of human eosinophils from nasal polyps reveals eosinophil heterogeneity in chronic rhinosinusitis tissue
Background
Chronic rhinosinusitis with nasal polyps (CRSwNP) is characterized by type 2 inflammation in the United States, but the actual roles that eosinophils play in CRSwNP remain largely unclear.
Objective
To reveal the roles and heterogeneity of eosinophils in nasal polyp (NP) tissue, we performed single cell RNA sequencing (scRNA-Seq) analysis of NP tissue.
Methods
Sinonasal tissues (NP and control sinus tissue) and patient matched peripheral blood (PB) samples were obtained from 5 control patients and 5 patients with CRSwNP. Eosinophils were enriched before processing for scRNA-Seq. The gene expression profiles in eosinophils were determined by microwell-based scRNA-Seq technology (BD Rhapsody platform). We predicted the overall function of NP eosinophils by Gene Ontology (geneontology.org) enrichment and pathway analyses and confirmed expression of selected genes by flow cytometry.
Results
After filtering out contaminating cells, we detected 5,542 eosinophils from control PB, 3,883 eosinophils from CRSwNP PB, 101 eosinophils from control sinus tissues (not included in further analyses), and 9,727 eosinophils from NPs by scRNA-Seq. We found that 204 genes were downregulated and 354 genes upregulated in NP eosinophils compared to all PB eosinophils (>1.5-fold, Padj < .05). Upregulated genes in NP eosinophils were associated with activation, cytokine-mediated signaling, growth factor activity, NF-κB signaling, and antiapoptotic molecules. NP eosinophils displayed 4 clusters revealing potential heterogeneity of eosinophils in NP tissue.
Conclusions
Elevated eosinophils in NP tissue appear to exist in several subtypes that may play important pathogenic roles in CRSwNP, in part by controlling inflammation and hyperproliferation of other cells.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
25.90
自引率
7.70%
发文量
1302
审稿时长
38 days
期刊介绍:
The Journal of Allergy and Clinical Immunology is a prestigious publication that features groundbreaking research in the fields of Allergy, Asthma, and Immunology. This influential journal publishes high-impact research papers that explore various topics, including asthma, food allergy, allergic rhinitis, atopic dermatitis, primary immune deficiencies, occupational and environmental allergy, and other allergic and immunologic diseases. The articles not only report on clinical trials and mechanistic studies but also provide insights into novel therapies, underlying mechanisms, and important discoveries that contribute to our understanding of these diseases. By sharing this valuable information, the journal aims to enhance the diagnosis and management of patients in the future.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


