对鼻拭子进行 MALDI-TOF MS 分析以确定 SARS-CoV-2 Omicron 感染者的特征
IF 9.7
4区 医学
Q1 MATERIALS SCIENCE, BIOMATERIALS
引用次数: 0
摘要
随着严重急性呼吸系统综合征冠状病毒 2(SARS-CoV-2)不断发生变异,导致各种变种,因此迫切需要新的 SARS-CoV-2 感染诊断方法。现有的核酸检测和抗原检测分别存在检测时间长和灵敏度低的问题。基于基质辅助激光解吸电离飞行时间质谱(MALDI-TOF MS)的鼻拭子分析已被证明是一种很有前途的 SARS-CoV-2 感染筛查技术。然而,该技术是否适用于 SARS-CoV-2 的不同变种尚不确定。鉴于自 2022 年以来 Omicron 变种的流行,我们开发了一种基于 MALDI-TOF 的鼻拭子样本诊断方法,以检测该变种的感染情况。我们采集了 325 份 SARS-CoV-2 阳性和 221 份 SARS-CoV-2 阴性鼻拭子样本,并通过 MALDI-TOF MS 获得了样本的分子质量指纹。使用随机森林机器学习分类模型分析分子质量指纹 MALDI-TOF 质谱,诊断 SARS-CoV-2 感染的准确率达到 97%,假阴性率为 0%,假阳性率为 7.6%。将 MALDI-TOF 分析与自上而下的蛋白质组学相结合,我们在鼻拭子中鉴定出了四个潜在的蛋白质生物标志物,即类人参素 4、胸腺肽 beta-10、胸腺肽 beta-4 和 statherin,用于 2019 年冠状病毒病的诊断。研究进一步发现,这四种蛋白质生物标志物还能区分 SARS-CoV-2 原始株感染和 Omicron 株感染。这些结果表明,基于MALDI-TOF MS的鼻拭子分析具有有效诊断SARS-CoV-2感染的能力,并具有全球应用和推广到其他传染病的潜力。本文章由计算机程序翻译,如有差异,请以英文原文为准。
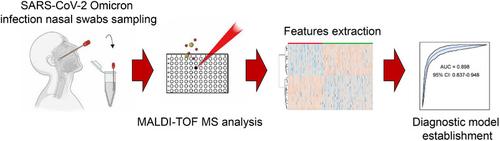
MALDI‐TOF MS analysis of nasal swabs for the characterization of patients infected with SARS‐CoV‐2 Omicron
With the ongoing mutation of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) leading to various variants, there is an urgent need for new diagnostic methods for SARS‐CoV‐2 infection. The existing nucleic acid test and antigen test suffer from long assay time and low sensitivity, respectively. Matrix‐assisted laser desorption/ionization time‐of‐flight mass spectrometry (MALDI‐TOF MS)‐based nasal swabs analysis have been demonstrated as a promising technique in SARS‐CoV‐2 infection screening. However, the applicability of the technique in the different variants of SARS‐CoV‐2 is uncertain. Given the prevalence of the Omicron variant since 2022, we developed a MALDI‐TOF‐based diagnosis method with nasal swab samples to detect the infection by this variant. We collected 325 SARS‐CoV‐2‐positive and 221 SARS‐CoV‐2‐negative nasal swab samples, and the molecular mass fingerprints were acquired from the samples by MALDI‐TOF MS. Using a random forest machine learning classification model to analyze the molecular mass fingerprints MALDI‐TOF mass spectra, the accuracy of 97%, false negative rate of 0%, and false positive rate of 7.6% were achieved for the diagnosis of SARS‐CoV‐2 infection. Combining the MALDI‐TOF analysis with top‐down proteomics, we identified four potential protein biomarkers, that is, humanin‐like 4, thymosin beta‐10, thymosin beta‐4 and statherin, in the nasal swab for the diagnosis of coronavirus disease 2019. It was further found that the four protein biomarkers can also differentiate the SARS‐CoV‐2 original strains infection and Omicron strains infection. These results suggest that the MALDI‐TOF MS‐based nasal swab analysis holds effective diagnostic capabilities of SARS‐CoV‐2 infection, and shows promising potential for global application and extension to other infectious diseases.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

VIEW
Multiple-
CiteScore
12.60
自引率
2.30%
发文量
0
审稿时长
10 weeks
期刊介绍:
View publishes scientific articles studying novel crucial contributions in the areas of Biomaterials and General Chemistry. View features original academic papers which go through peer review by experts in the given subject area.View encourages submissions from the research community where the priority will be on the originality and the practical impact of the reported research.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


