高通量鉴定肠道微生物依赖性代谢物。
IF 13.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
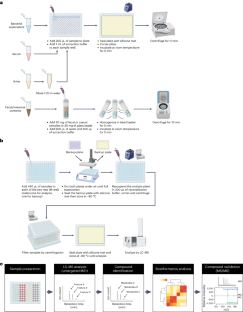
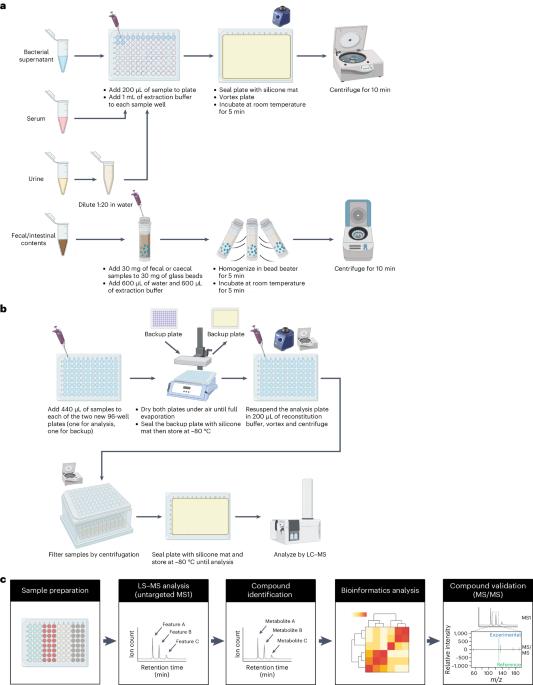
鉴定和研究肠道微生物产生的各种代谢物是限制微生物组科学进步的一个重要障碍。肠道微生物代谢产生数千种难以识别的代谢物,这给研究它们在宿主生物学中的作用带来了挑战。近年来,基于质谱的代谢组学已成为鉴定小分子代谢物的核心技术之一。然而,学术实验室往往缺乏从样品制备到仪器使用和数据分析的代谢组学专业知识。大多数靶向代谢组学方法都能提供高灵敏度和高定量水平,但它们仅限于一组预定义的分子,对以微生物组为重点的研究可能没有参考价值。在这里,我们利用液相色谱-质谱联用技术和生物信息分析技术开发了一种以肠道微生物为重点的广谱代谢组学方案。用户只需使用液相色谱-质谱联用仪,就能完成从样品采集到数据分析的整个实验过程。通过将该方案应用于含有人类肠道微生物代谢物的样本(从培养上清液到人类生物样本),我们的方法可以高置信度地鉴定 >800 种代谢物,这些代谢物可作为微生物-宿主相互作用的候选介质。我们希望该方案能降低体外和哺乳动物宿主体内追踪肠道细菌代谢的障碍,推动假设驱动的机理研究,加快我们对肠道微生物组化学水平的了解。本文章由计算机程序翻译,如有差异,请以英文原文为准。
High-throughput identification of gut microbiome-dependent metabolites
A significant hurdle that has limited progress in microbiome science has been identifying and studying the diverse set of metabolites produced by gut microbes. Gut microbial metabolism produces thousands of difficult-to-identify metabolites, which present a challenge to study their roles in host biology. In recent years, mass spectrometry-based metabolomics has become one of the core technologies for identifying small metabolites. However, metabolomics expertise, ranging from sample preparation to instrument use and data analysis, is often lacking in academic labs. Most targeted metabolomics methods provide high levels of sensitivity and quantification, while they are limited to a panel of predefined molecules that may not be informative to microbiome-focused studies. Here we have developed a gut microbe-focused and wide-spectrum metabolomic protocol using liquid chromatography–mass spectrometry and bioinformatic analysis. This protocol enables users to carry out experiments from sample collection to data analysis, only requiring access to a liquid chromatography–mass spectrometry instrument, which is often available at local core facilities. By applying this protocol to samples containing human gut microbial metabolites, spanning from culture supernatant to human biospecimens, our approach enables high-confidence identification of >800 metabolites that can serve as candidate mediators of microbe–host interactions. We expect this protocol will lower the barrier to tracking gut bacterial metabolism in vitro and in mammalian hosts, propelling hypothesis-driven mechanistic studies and accelerating our understanding of the gut microbiome at the chemical level. This protocol presents a metabolomics method tailored for detecting and measuring gut-microbe-derived metabolites using a broad reference library of metabolite standards.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Protocols
生物-生化研究方法
CiteScore
29.10
自引率
0.70%
发文量
128
审稿时长
4 months
期刊介绍:
Nature Protocols focuses on publishing protocols used to address significant biological and biomedical science research questions, including methods grounded in physics and chemistry with practical applications to biological problems. The journal caters to a primary audience of research scientists and, as such, exclusively publishes protocols with research applications. Protocols primarily aimed at influencing patient management and treatment decisions are not featured.
The specific techniques covered encompass a wide range, including but not limited to: Biochemistry, Cell biology, Cell culture, Chemical modification, Computational biology, Developmental biology, Epigenomics, Genetic analysis, Genetic modification, Genomics, Imaging, Immunology, Isolation, purification, and separation, Lipidomics, Metabolomics, Microbiology, Model organisms, Nanotechnology, Neuroscience, Nucleic-acid-based molecular biology, Pharmacology, Plant biology, Protein analysis, Proteomics, Spectroscopy, Structural biology, Synthetic chemistry, Tissue culture, Toxicology, and Virology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


