TAC-TIC,一种用于识别细菌毒素-抗毒素系统触发器或阻断器的高通量遗传学方法。
IF 13.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
毒素-抗毒素系统(TAs)在细菌染色体中含量丰富,可在压力下阻止生长,但通常保持非活性。越来越多的人认为,TAs 可使受噬菌体或外来遗传因子1,2 感染的细菌停止生长,从而保护种群(中止感染,Abi)。TAs和其他Abi系统3的多样性和丰富性表明它们发挥着重要的免疫作用,然而是什么让它们能够感知攻击,这在很大程度上仍然是个谜。在这里,我们介绍了一种名为毒素激活-抑制共轭(TAC-TIC)的方法,用来鉴定触发或阻断噬菌体防御三方再创-TAs毒性的基因产物4。TAC-TIC 采用高密度阵列可移动基因表达库,通过可诱导载体将其转移到携带完整 TA 系统或仅携带其毒性成分的细胞中。然后将双质粒转染体固定在含诱导剂的琼脂平板上,并对其菌落适应性进行量化,以确定触发 TA 抑制生长(TAC)或阻止 TA 起作用(TIC)的基因产物。TAC-TIC针对辛格ROTOR钉螺机器人进行了优化,但也可用于其他机器人或人工钉螺,可在一周内筛选出数万个针对任何TA或Abi(具有毒性)的基因。最后,我们介绍了一种双共轭供体/克隆菌株(大肠杆菌 DATC),它能加速从噬菌体构建 TAC-TIC 基因供体文库,从而利用 TAC-TIC 鉴定噬菌体基因组中的 TA 触发器和抗防御机制。本文章由计算机程序翻译,如有差异,请以英文原文为准。
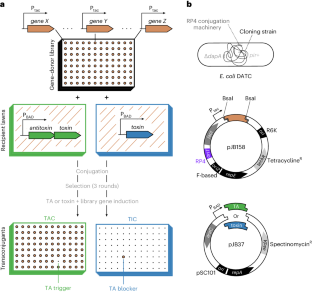
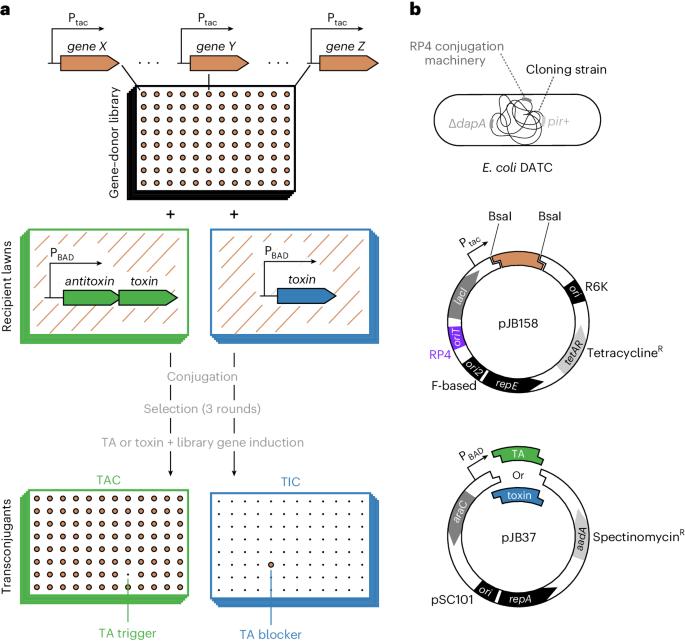
TAC–TIC, a high-throughput genetics method to identify triggers or blockers of bacterial toxin–antitoxin systems
Toxin–antitoxin systems (TAs) are abundant in bacterial chromosomes and can arrest growth under stress, but usually remain inactive. TAs have been increasingly implicated in halting the growth of infected bacteria from bacteriophages or foreign genetic elements1,2 to protect the population (abortive infection, Abi). The vast diversity and abundance of TAs and other Abi systems3 suggest they play an important immunity role, yet what allows them to sense attack remains largely enigmatic. Here, we describe a method called toxin activation–inhibition conjugation (TAC–TIC), which we used to identify gene products that trigger or block the toxicity of phage-defending tripartite retron-TAs4. TAC–TIC employs high-density arrayed mobilizable gene-overexpression libraries, which are transferred into cells carrying the full TA system or only its toxic component, on inducible vectors. The double-plasmid transconjugants are then pinned on inducer-containing agar plates and their colony fitness is quantified to identify gene products that trigger a TA to inhibit growth (TAC), or that block it from acting (TIC). TAC–TIC is optimized for the Singer ROTOR pinning robot, but can also be used with other robots or manual pinners, and allows screening tens of thousands of genes against any TA or Abi (with toxicity) within a week. Finally, we present a dual conjugation donor/cloning strain (Escherichia coli DATC), which accelerates the construction of TAC–TIC gene-donor libraries from phages, enabling the use of TAC–TIC for identifying TA triggers and antidefense mechanisms in phage genomes. This protocol describes toxin activation–inhibition conjugation (TAC–TIC), a reverse genetics screening approach that can be used to identify triggers or blockers of bacterial toxin–antitoxin or phage immunity systems.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Protocols
生物-生化研究方法
CiteScore
29.10
自引率
0.70%
发文量
128
审稿时长
4 months
期刊介绍:
Nature Protocols focuses on publishing protocols used to address significant biological and biomedical science research questions, including methods grounded in physics and chemistry with practical applications to biological problems. The journal caters to a primary audience of research scientists and, as such, exclusively publishes protocols with research applications. Protocols primarily aimed at influencing patient management and treatment decisions are not featured.
The specific techniques covered encompass a wide range, including but not limited to: Biochemistry, Cell biology, Cell culture, Chemical modification, Computational biology, Developmental biology, Epigenomics, Genetic analysis, Genetic modification, Genomics, Imaging, Immunology, Isolation, purification, and separation, Lipidomics, Metabolomics, Microbiology, Model organisms, Nanotechnology, Neuroscience, Nucleic-acid-based molecular biology, Pharmacology, Plant biology, Protein analysis, Proteomics, Spectroscopy, Structural biology, Synthetic chemistry, Tissue culture, Toxicology, and Virology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


