驱动抗菌药耐药性在患者内部产生的生态和进化机制
IF 69.2
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
摘要
人们对抗菌药耐药性(AMR)在患者体内产生的生态和进化机制以及这些机制在不同细菌感染中的差异知之甚少。病原体基因组测序技术在临床中的应用日益广泛,使我们能够更深入地了解这些过程。在本综述中,我们将探讨临床证据支持细菌在患者体内出现抗药性的四种主要机制:自发抗药性突变;抗药性基因的原位水平基因转移;原有抗药性的选择;以及抗药性菌系的迁移。患者体内的 AMR 出现在多种宿主生态位和细菌种类中,但每种机制的重要性因细菌种类和体内感染部位而异。我们确定了这种差异的潜在驱动因素,并讨论了如何将生态和进化分析嵌入抗菌药物临床试验中,这些试验是了解这些机制为何在病原体、感染和个体之间存在差异的有力工具,但却未得到充分利用。最终,如果能更好地了解宿主生态位、细菌种类和抗生素作用模式如何共同作用于患者体内 AMR 出现的生态和进化机制,就能实现更具预测性和个性化的诊断和抗菌疗法。本文章由计算机程序翻译,如有差异,请以英文原文为准。
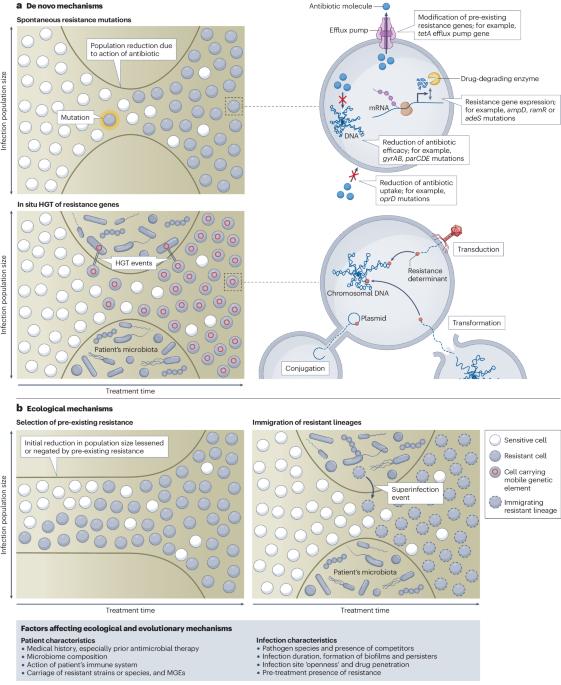
Ecological and evolutionary mechanisms driving within-patient emergence of antimicrobial resistance
The ecological and evolutionary mechanisms of antimicrobial resistance (AMR) emergence within patients and how these vary across bacterial infections are poorly understood. Increasingly widespread use of pathogen genome sequencing in the clinic enables a deeper understanding of these processes. In this Review, we explore the clinical evidence to support four major mechanisms of within-patient AMR emergence in bacteria: spontaneous resistance mutations; in situ horizontal gene transfer of resistance genes; selection of pre-existing resistance; and immigration of resistant lineages. Within-patient AMR emergence occurs across a wide range of host niches and bacterial species, but the importance of each mechanism varies between bacterial species and infection sites within the body. We identify potential drivers of such differences and discuss how ecological and evolutionary analysis could be embedded within clinical trials of antimicrobials, which are powerful but underused tools for understanding why these mechanisms vary between pathogens, infections and individuals. Ultimately, improving understanding of how host niche, bacterial species and antibiotic mode of action combine to govern the ecological and evolutionary mechanism of AMR emergence in patients will enable more predictive and personalized diagnosis and antimicrobial therapies. In this Review, Shepherd, Brockhurst and colleagues explore the clinical evidence in support of four major ecological and evolutionary mechanisms of within-patient antimicrobial resistance emergence in bacteria, and how host niche, bacterial species and antibiotic mode of action combine to govern these mechanisms.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Nature Reviews Microbiology
生物-微生物学
CiteScore
74.00
自引率
0.50%
发文量
149
审稿时长
6-12 weeks
期刊介绍:
At Nature Reviews Microbiology, our goal is to become the leading source of reviews and commentaries for the scientific community we cater to. We are dedicated to publishing articles that are not only authoritative but also easily accessible, supplementing them with clear and concise figures, tables, and other visual aids. Our objective is to offer an unparalleled service to authors, referees, and readers, and we continuously strive to maximize the usefulness and impact of each article we publish. With a focus on Reviews, Perspectives, and Comments spanning the entire field of microbiology, our wide scope ensures that the work we feature reaches the widest possible audience.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


