人类外显子定义剪接体激活前的结构研究。
IF 28.1
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
摘要
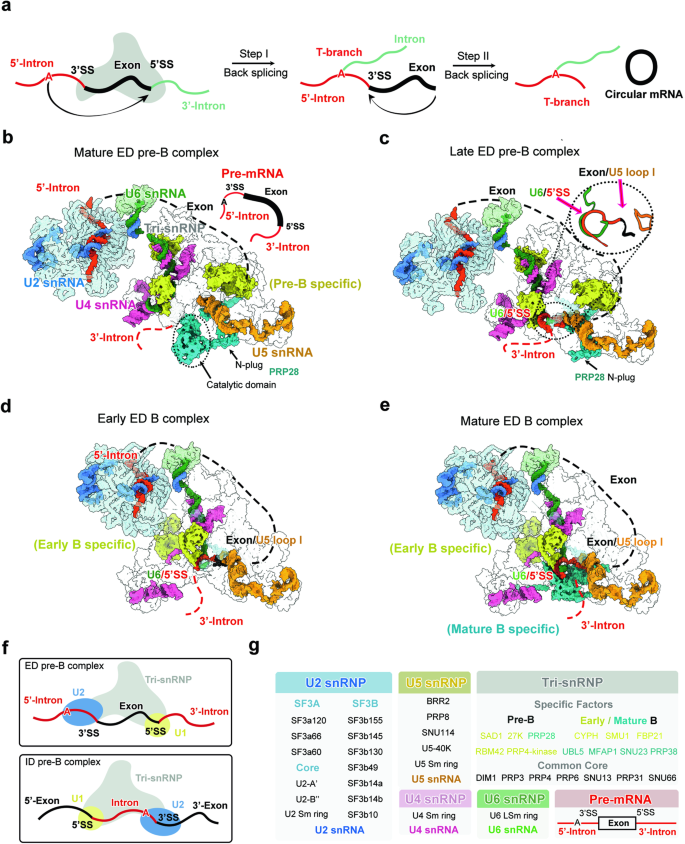
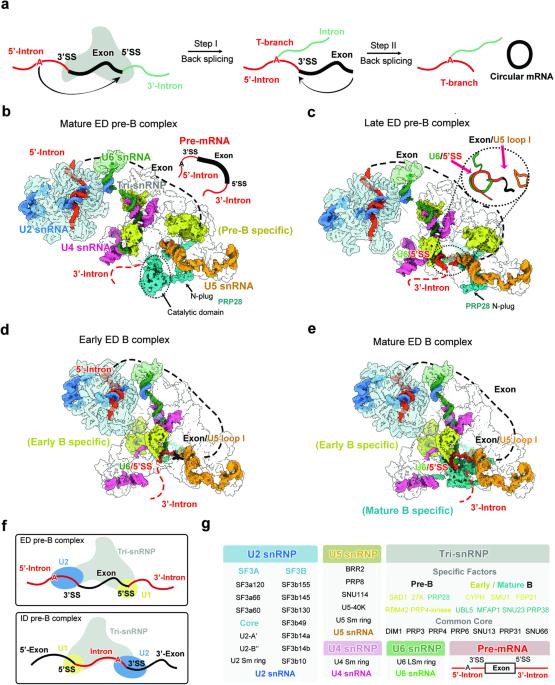
剪接体通常在一个外显子上组装,然后经过重排跨越邻近的内含子。内含子定义的剪接体的大多数状态已在结构上得到表征。然而,完全组装的外显子定义剪接体的结构仍未确定。在剪接体组装过程中,前催化状态(B 复合物)由其前体(pre-B 复合物)转化而来。在先前未知的晚期前 B 状态中,U1 snRNP 已经释放,但其余蛋白质仍处于前 B 状态;意外的是,RNA 处于 B 状态,U6 snRNA 与 5′-剪接位点形成双链,U5 snRNA 识别外显子的 3′-末端。在早期和成熟的 B 复合物中,B 特异因子被逐步招募,并特异性地识别外显子 3′区。我们的研究揭示了外显子定义的剪接体的组装过程,并确定了前 B 向 B 过渡的机制步骤。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Structural insights into human exon-defined spliceosome prior to activation
Spliceosome is often assembled across an exon and undergoes rearrangement to span a neighboring intron. Most states of the intron-defined spliceosome have been structurally characterized. However, the structure of a fully assembled exon-defined spliceosome remains at large. During spliceosome assembly, the pre-catalytic state (B complex) is converted from its precursor (pre-B complex). Here we report atomic structures of the exon-defined human spliceosome in four sequential states: mature pre-B, late pre-B, early B, and mature B. In the previously unknown late pre-B state, U1 snRNP is already released but the remaining proteins are still in the pre-B state; unexpectedly, the RNAs are in the B state, with U6 snRNA forming a duplex with 5′-splice site and U5 snRNA recognizing the 3′-end of the exon. In the early and mature B complexes, the B-specific factors are stepwise recruited and specifically recognize the exon 3′-region. Our study reveals key insights into the assembly of the exon-defined spliceosomes and identifies mechanistic steps of the pre-B-to-B transition.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cell Research
生物-细胞生物学
CiteScore
53.90
自引率
0.70%
发文量
2420
审稿时长
2.3 months
期刊介绍:
Cell Research (CR) is an international journal published by Springer Nature in partnership with the Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences (CAS). It focuses on publishing original research articles and reviews in various areas of life sciences, particularly those related to molecular and cell biology. The journal covers a broad range of topics including cell growth, differentiation, and apoptosis; signal transduction; stem cell biology and development; chromatin, epigenetics, and transcription; RNA biology; structural and molecular biology; cancer biology and metabolism; immunity and molecular pathogenesis; molecular and cellular neuroscience; plant molecular and cell biology; and omics, system biology, and synthetic biology. CR is recognized as China's best international journal in life sciences and is part of Springer Nature's prestigious family of Molecular Cell Biology journals.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


