基于荧光素酶和 HaloTag 的报告分析法测量活细胞中小分子诱导的降解途径
IF 13.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
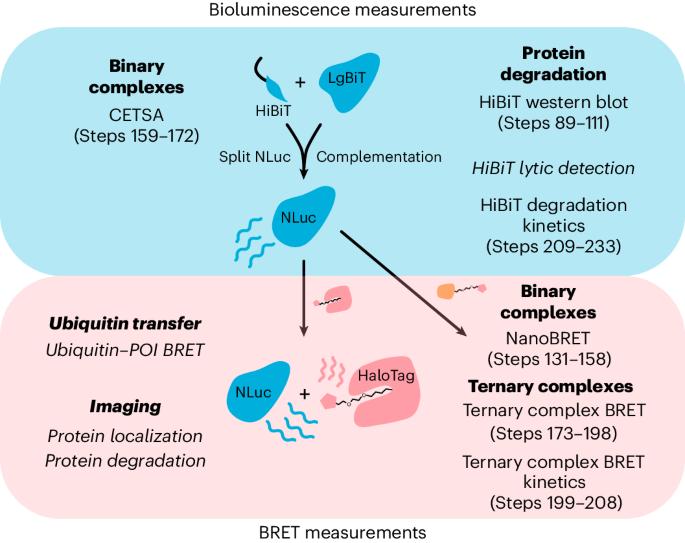
小分子降解剂(如蛋白水解靶向嵌合体)的合理开发仍然是一项挑战,因为决定降解剂效率的限速步骤在很大程度上是未知的。靶向蛋白质降解领域的标准方法大多依赖于经典的低通量终点检测,如 Western 印迹或定量蛋白质组学。在这里,我们应用了基于纳米荧光素酶和 HaloTag 的筛选技术,以确定小分子诱导的相关蛋白质与选定的 E3 连接酶之间形成三元复合物的动力学和稳定性。我们设计了一系列活细胞检测方法来探测降解过程中最关键的步骤,同时最大限度地减少所需的表达构建体的数量,从而使所提出的检测管道具有灵活性并能适应用户的要求。这种方法可以评估选择性目标降解剂的基本机制,并揭示所开发的降解剂分子在活细胞中的确切特性。该方案使受过基础细胞培养和分子生物学培训的科学家能够在 2 周内通过跟踪三元复合物的形成进行小分子近端诱导剂筛选,而使用 HiBiT 系统进行降解剂筛选则需要 CRISPR-Cas9 工程细胞系,其生成时间可能长达 3 个月。细胞系生成后,可在数天内以高通量方式进行降解剂筛选和验证。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Luciferase- and HaloTag-based reporter assays to measure small-molecule-induced degradation pathway in living cells
The rational development of small-molecule degraders (e.g., proteolysis targeting chimeras) remains a challenge as the rate-limiting steps that determine degrader efficiency are largely unknown. Standard methods in the field of targeted protein degradation mostly rely on classical, low-throughput endpoint assays such as western blots or quantitative proteomics. Here we applied NanoLuciferase- and HaloTag-based screening technologies to determine the kinetics and stability of small-molecule-induced ternary complex formation between a protein of interest and a selected E3 ligase. A collection of live-cell assays were designed to probe the most critical steps of the degradation process while minimizing the number of required expression constructs, making the proposed assay pipeline flexible and adaptable to the requirements of the users. This approach evaluates the underlying mechanism of selective target degraders and reveals the exact characteristics of the developed degrader molecules in living cells. The protocol allows scientists trained in basic cell culture and molecular biology to carry out small-molecule proximity-inducer screening via tracking of the ternary complex formation within 2 weeks of establishment, while degrader screening using the HiBiT system requires a CRISPR–Cas9 engineered cell line whose generation can take up to 3 months. After cell-line generation, degrader screening and validation can be carried out in high-throughput manner within days. NanoLuciferase- and HaloTag-based screening technologies are versatile tools suitable for the live-cell analysis of the entire small-molecule-induced degradation cascade to uncover the mode of action of proximity-inducing compounds such as PROTACs.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Protocols
生物-生化研究方法
CiteScore
29.10
自引率
0.70%
发文量
128
审稿时长
4 months
期刊介绍:
Nature Protocols focuses on publishing protocols used to address significant biological and biomedical science research questions, including methods grounded in physics and chemistry with practical applications to biological problems. The journal caters to a primary audience of research scientists and, as such, exclusively publishes protocols with research applications. Protocols primarily aimed at influencing patient management and treatment decisions are not featured.
The specific techniques covered encompass a wide range, including but not limited to: Biochemistry, Cell biology, Cell culture, Chemical modification, Computational biology, Developmental biology, Epigenomics, Genetic analysis, Genetic modification, Genomics, Imaging, Immunology, Isolation, purification, and separation, Lipidomics, Metabolomics, Microbiology, Model organisms, Nanotechnology, Neuroscience, Nucleic-acid-based molecular biology, Pharmacology, Plant biology, Protein analysis, Proteomics, Spectroscopy, Structural biology, Synthetic chemistry, Tissue culture, Toxicology, and Virology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


