HS-AFM 单分子结构生物学揭示了转运体徘徊动力学的基础
IF 12.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
Pyrococcus horikoshii 的氨基酸转运体 GltPh 与其他通道和转运体一样,具有活动模式切换功能,即以前所说的徘徊动力学。遗憾的是,到目前为止,人们还不了解这些活动波动的基础,这可能是由于缺乏直接获取与转运体瞬时活动相关的结构特征的实验工具。在这里,我们利用高速原子力显微镜(其独特之处在于可同时提供结构和时间分辨率)来揭示蛋白质动力学模式切换的基础。我们开发了膜延伸膜蛋白重组技术,可对分离的分子进行分析。结合定位原子力显微镜、主成分分析和隐马尔可夫模型,我们可以将结构状态与功能时间线联系起来,从而可以从单个分子中解析出六种结构,并确定了一种内向状态(IFSopen-1),作为构象景观中的动力学死胡同。在 GltPh 上介绍的方法普遍适用,为时间分辨动态单分子结构生物学开辟了可能性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
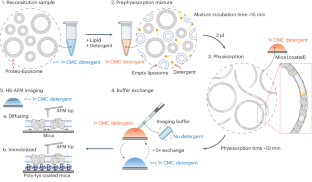
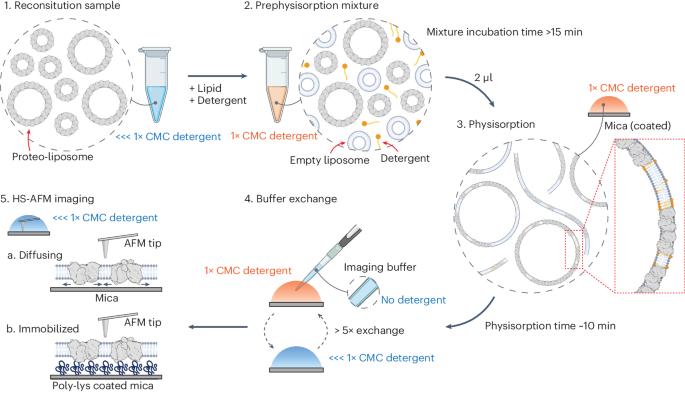
HS-AFM single-molecule structural biology uncovers basis of transporter wanderlust kinetics
The Pyrococcus horikoshii amino acid transporter GltPh revealed, like other channels and transporters, activity mode switching, previously termed wanderlust kinetics. Unfortunately, to date, the basis of these activity fluctuations is not understood, probably due to a lack of experimental tools that directly access the structural features of transporters related to their instantaneous activity. Here, we take advantage of high-speed atomic force microscopy, unique in providing simultaneous structural and temporal resolution, to uncover the basis of kinetic mode switching in proteins. We developed membrane extension membrane protein reconstitution that allows the analysis of isolated molecules. Together with localization atomic force microscopy, principal component analysis and hidden Markov modeling, we could associate structural states to a functional timeline, allowing six structures to be solved from a single molecule, and an inward-facing state, IFSopen-1, to be determined as a kinetic dead-end in the conformational landscape. The approaches presented on GltPh are generally applicable and open possibilities for time-resolved dynamic single-molecule structural biology. Combining high-speed atomic force microscopy (AFM) with localization AFM and principal component analysis, the authors present six structures of a glutamate transporter and associate the conformational states to the molecule’s activity timeline.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Structural & Molecular Biology
BIOCHEMISTRY & MOLECULAR BIOLOGY-BIOPHYSICS
CiteScore
22.00
自引率
1.80%
发文量
160
审稿时长
3-8 weeks
期刊介绍:
Nature Structural & Molecular Biology is a comprehensive platform that combines structural and molecular research. Our journal focuses on exploring the functional and mechanistic aspects of biological processes, emphasizing how molecular components collaborate to achieve a particular function. While structural data can shed light on these insights, our publication does not require them as a prerequisite.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


