经历瓶颈和增长的亚种群复杂性状的遗传率
IF 2.6
3区 生物学
Q2 GENETICS & HEREDITY
引用次数: 0
摘要
全基因组关联研究(GWAS)经常使用经历过瓶颈期的人群来研究与复杂性状相关的变异。一般认为,这些与世隔绝的亚人群可能会出现高频率、大效应的罕见变异,因此为研究上述性状提供了独特的机会。然而,被研究人群的人口历史会影响决定复杂性状全基因组的所有 SNPs,从而改变其遗传率和遗传结构。我们采用一种基于模拟的方法来确定漂移、扩张和迁移等人口统计过程对复杂性状遗传率的影响。我们表明,人口统计学对复杂性状有相当大的影响。然后,我们研究了在受人口统计学影响的 GWAS 研究中解析复杂性状遗传率的能力。我们发现,人口统计学是解释复杂性状推论的一个重要组成部分,对 GWAS 的能力有细微的影响。我们的结论是,需要对人口统计历史进行明确建模,以正确量化复杂性状的选择历史。本文章由计算机程序翻译,如有差异,请以英文原文为准。
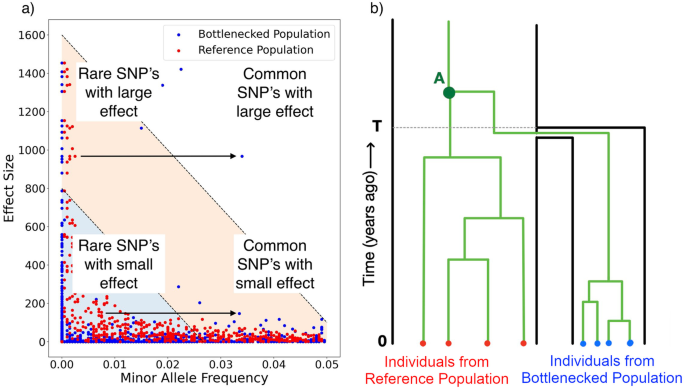
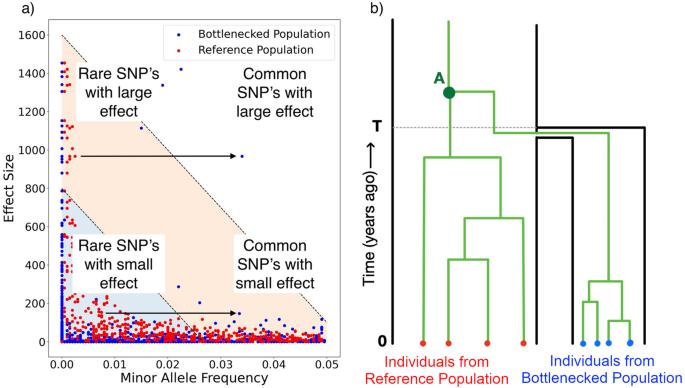
Heritability of complex traits in sub-populations experiencing bottlenecks and growth
Populations that have experienced a bottleneck are regularly used in Genome Wide Association Studies (GWAS) to investigate variants associated with complex traits. It is generally understood that these isolated sub-populations may experience high frequency of otherwise rare variants with large effect size, and therefore provide a unique opportunity to study said trait. However, the demographic history of the population under investigation affects all SNPs that determine the complex trait genome-wide, changing its heritability and genetic architecture. We use a simulation based approach to identify the impact of the demographic processes of drift, expansion, and migration on the heritability of complex trait. We show that demography has considerable impact on complex traits. We then investigate the power to resolve heritability of complex traits in GWAS studies subjected to demographic effects. We find that demography is an important component for interpreting inference of complex traits and has a nuanced impact on the power of GWAS. We conclude that demographic histories need to be explicitly modelled to properly quantify the history of selection on a complex trait.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Human Genetics
生物-遗传学
CiteScore
7.20
自引率
0.00%
发文量
101
审稿时长
4-8 weeks
期刊介绍:
The Journal of Human Genetics is an international journal publishing articles on human genetics, including medical genetics and human genome analysis. It covers all aspects of human genetics, including molecular genetics, clinical genetics, behavioral genetics, immunogenetics, pharmacogenomics, population genetics, functional genomics, epigenetics, genetic counseling and gene therapy.
Articles on the following areas are especially welcome: genetic factors of monogenic and complex disorders, genome-wide association studies, genetic epidemiology, cancer genetics, personal genomics, genotype-phenotype relationships and genome diversity.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


