基因组比较揭示了对苹果球孢霉叶斑病致病性进化至关重要的附属基因
IF 4.8
1区 农林科学
Q1 PLANT SCIENCES
引用次数: 0
摘要
苹果球孢菌叶斑病(GLS)是一种新出现的真菌病害,由 Colletotrichum fructicola 和其他 Colletotrichum 菌种引起。这些物种是多态的,目前还不清楚这些病原体是如何进化到感染苹果的。我们利用长线程测序技术生成了果实疫霉菌中一个适应 GLS 的分离株和一个不适应 GLS 的分离株的染色体组。此外,我们还利用短线程测序技术对 17 个果蝇科 C. 和 C. aenigma 分离物进行了重新测序,这些分离物的 GLS 致病性各不相同。基因组比较显示,果孢子菌和球孢子菌种群中其他密切相关的菌种具有保守的两部分基因组结构,其中包括小染色体(附属染色体)。此外,在 C. fructicola 和 C. aenigma 的 GLS 致病分离株中,有两个重复丰富的基因组区域(共 1.61 Mb)是特别保守的。在 C. fructicola 的 GLS 特异性区域内对 10 个附属基因进行了单基因缺失,发现其中 3 个基因对 GLS 的致病性至关重要。这些基因编码一种假定的非核糖体肽合成酶、一种黄素结合单氧化酶和一种功能未知的小蛋白。这些结果凸显了附属基因在 Colletotrichum 致病性进化过程中发挥的关键作用,并暗示了一种未确定的次生代谢物在 GLS 致病过程中的重要性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
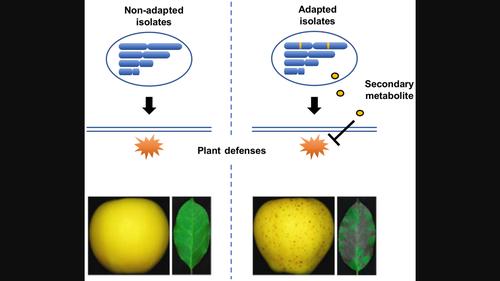
Genome comparisons reveal accessory genes crucial for the evolution of apple Glomerella leaf spot pathogenicity in Colletotrichum fungi
Apple Glomerella leaf spot (GLS) is an emerging fungal disease caused by Colletotrichum fructicola and other Colletotrichum species. These species are polyphyletic and it is currently unknown how these pathogens convergently evolved to infect apple. We generated chromosome‐level genome assemblies of a GLS‐adapted isolate and a non‐adapted isolate in C. fructicola using long‐read sequencing. Additionally, we resequenced 17 C. fructicola and C. aenigma isolates varying in GLS pathogenicity using short‐read sequencing. Genome comparisons revealed a conserved bipartite genome architecture involving minichromosomes (accessory chromosomes) shared by C. fructicola and other closely related species within the C. gloeosporioides species complex. Moreover, two repeat‐rich genomic regions (1.61 Mb in total) were specifically conserved among GLS‐pathogenic isolates in C. fructicola and C. aenigma . Single‐gene deletion of 10 accessory genes within the GLS‐specific regions of C. fructicola identified three that were essential for GLS pathogenicity. These genes encoded a putative non‐ribosomal peptide synthetase, a flavin‐binding monooxygenase and a small protein with unknown function. These results highlight the crucial role accessory genes play in the evolution of Colletotrichum pathogenicity and imply the significance of an unidentified secondary metabolite in GLS pathogenesis.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Molecular plant pathology
生物-植物科学
CiteScore
9.40
自引率
4.10%
发文量
120
审稿时长
6-12 weeks
期刊介绍:
Molecular Plant Pathology is now an open access journal. Authors pay an article processing charge to publish in the journal and all articles will be freely available to anyone. BSPP members will be granted a 20% discount on article charges. The Editorial focus and policy of the journal has not be changed and the editorial team will continue to apply the same rigorous standards of peer review and acceptance criteria.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


