黄瓜抗疫霉病候选基因的精细图谱绘制与分析
IF 3.6
3区 生物学
Q1 PLANT SCIENCES
引用次数: 0
摘要
黄瓜枯萎病是一种毁灭性病害。控制这种病害的最佳方法是抗病育种。本研究的重点是抗病基因图谱和分子标记的开发。我们以抗病黄瓜 JSH 和感病黄瓜 B80 为亲本,构建 F2 群体。通过大量分离分析(BSA)和特异长度扩增片段测序(SLAF-seq),我们开发了裂解扩增多态性序列(CAPs)标记来绘制抗病基因图谱。F2个体的抗性表现出抗性与易感性的分离比接近3:1。抗JSH黄瓜的基因被映射到一个9.25 kb的区间,映射区三个基因的测序结果显示,在JSH和B80之间的Csa5G139130编码区的225、302和591碱基位点有三个突变,但Csa5G139140和Csa5G139150编码区没有突变。突变导致 Csa5G139130 所编码蛋白质的第 75 和 101 个氨基酸发生变化,这表明 Csa5G139130 最有可能是抗性候选基因。我们开发了一个分子标记 CAPs-4,作为与黄瓜枯萎病抗性基因密切相关的标记。这是首次报道黄瓜抗枯萎病基因的图谱绘制,将为黄瓜抗枯萎病的分子育种提供有用的标记。本文章由计算机程序翻译,如有差异,请以英文原文为准。
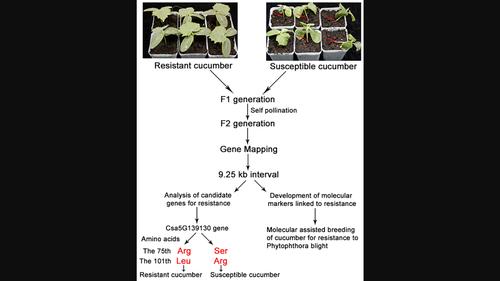
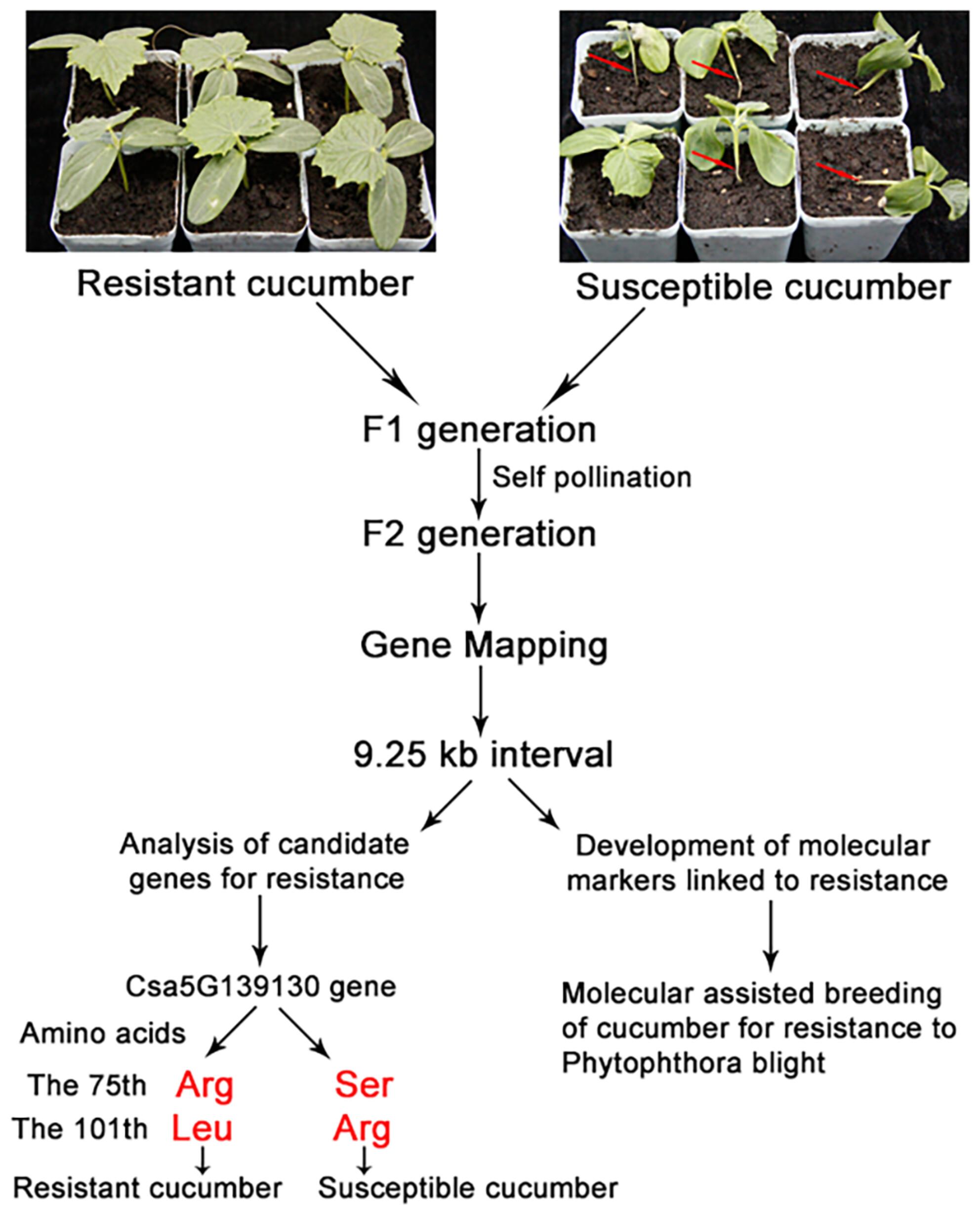
Fine mapping and analysis of a candidate gene controlling Phytophthora blight resistance in cucumber
- Cucumber blight is a destructive disease. The best way to control this disease is resistance breeding. This study focuses on disease resistance gene mapping and molecular marker development.
- We used the resistant cucumber, JSH, and susceptible cucumber, B80, as parents to construct F2 populations. Bulked segregant analysis (BSA) combined with specific length amplified fragment sequencing (SLAF-seq) were used, from which we developed cleaved amplified polymorphic sequence (CAPs) markers to map the resistance gene.
- Resistance in F2 individuals showed a segregation ratio of resistance:susceptibility close to 3:1. The gene in JSH resistant cucumber was mapped to an interval of 9.25 kb, and sequencing results for the three genes in the mapped region revealed three mutations at base sites 225, 302, and 591 in the coding region of Csa5G139130 between JSH and B80, but no mutations in coding regions of Csa5G139140 and Csa5G139150. The mutations caused changes in amino acids 75 and 101 of the protein encoded by Csa5G139130, suggesting that Csa5G139130 is the most likely resistance candidate gene. We developed a molecular marker, CAPs-4, as a closely linked marker for the cucumber blight resistance gene.
- This is the first report on mapping of a cucumber blight resistance gene and will provideg a useful marker for molecular breeding of cucumber resistance to Phytophthora blight.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Plant Biology
生物-植物科学
CiteScore
8.20
自引率
2.60%
发文量
109
审稿时长
3 months
期刊介绍:
Plant Biology is an international journal of broad scope bringing together the different subdisciplines, such as physiology, molecular biology, cell biology, development, genetics, systematics, ecology, evolution, ecophysiology, plant-microbe interactions, and mycology.
Plant Biology publishes original problem-oriented full-length research papers, short research papers, and review articles. Discussion of hot topics and provocative opinion articles are published under the heading Acute Views. From a multidisciplinary perspective, Plant Biology will provide a platform for publication, information and debate, encompassing all areas which fall within the scope of plant science.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


