通过大规模钙成像和机器学习揭示丘脑上核的系统级时间计算和表征
IF 28.1
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
摘要
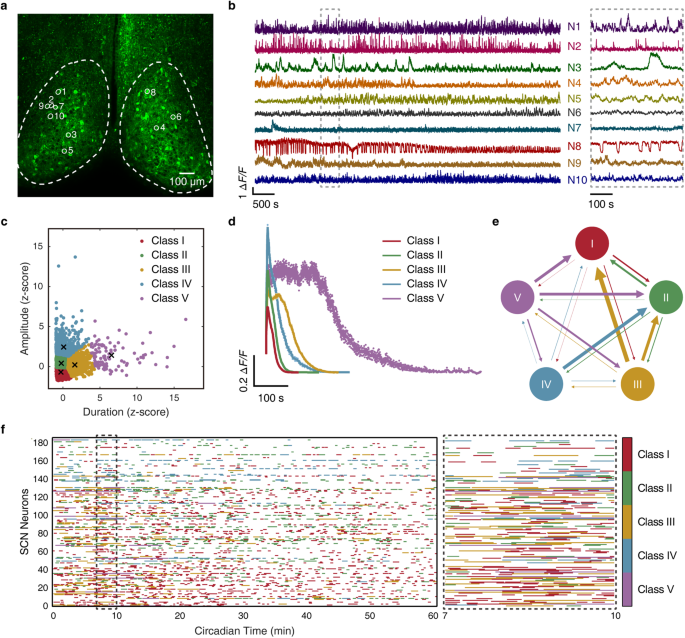
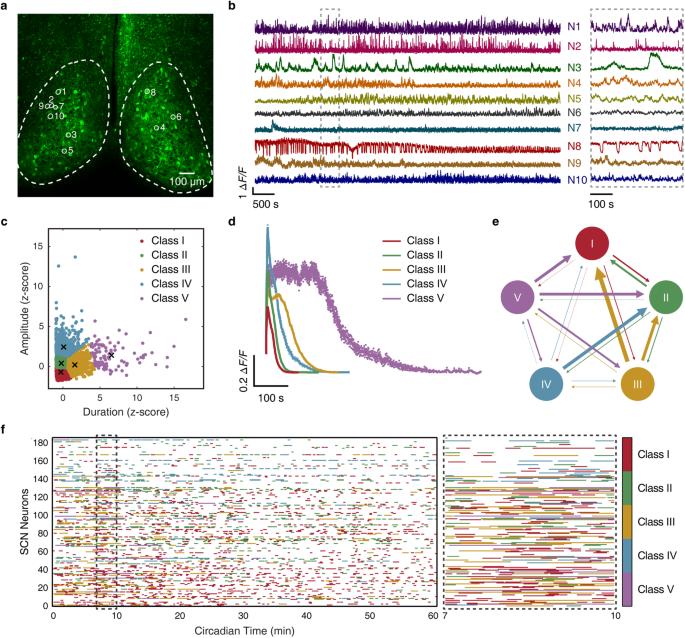
嗜铬细胞上核(SCN)是哺乳动物的中枢昼夜节律起搏器,其中的异质神经元协同作用,而每个神经元都有一个自我维持的分子钟表。然而,系统级 SCN 信号如何编码一天中的时间仍然是个谜。在这里,我们展示了种群级 Ca2+ 信号通过与 SCN 中的空间模块化时间特征表征相结合的群体决策机制来预测每小时的时间。具体来说,我们开发了一种高速双视角双光子显微镜,用于对成人 SCN 切片中多达 9000 个 GABA 能神经元进行容量 Ca2+ 成像,并利用机器学习方法从多尺度 Ca2+ 信号整体中捕捉突现特性。我们通过对 SCN 神经元随机群组进行轮询,实现了每小时时间预测,在群组规模为 900 个时,准确率达到 99.0%。此外,我们还发现,通过对比学习识别出的功能神经元亚型往往会在 SCN 空间中单独聚集,形成双侧对称的波纹状模块模式。单个模块代表了独特的时间特征,因此针对特定模块学习的时间预测器也能从同一模块的随机轮询中准确解码每小时的时间。这些发现为在系统水平上解读生物钟的设计原理开辟了新的范式。本文章由计算机程序翻译,如有差异,请以英文原文为准。
System-level time computation and representation in the suprachiasmatic nucleus revealed by large-scale calcium imaging and machine learning
The suprachiasmatic nucleus (SCN) is the mammalian central circadian pacemaker with heterogeneous neurons acting in concert while each neuron harbors a self-sustained molecular clockwork. Nevertheless, how system-level SCN signals encode time of the day remains enigmatic. Here we show that population-level Ca2+ signals predict hourly time, via a group decision-making mechanism coupled with a spatially modular time feature representation in the SCN. Specifically, we developed a high-speed dual-view two-photon microscope for volumetric Ca2+ imaging of up to 9000 GABAergic neurons in adult SCN slices, and leveraged machine learning methods to capture emergent properties from multiscale Ca2+ signals as a whole. We achieved hourly time prediction by polling random cohorts of SCN neurons, reaching 99.0% accuracy at a cohort size of 900. Further, we revealed that functional neuron subtypes identified by contrastive learning tend to aggregate separately in the SCN space, giving rise to bilaterally symmetrical ripple-like modular patterns. Individual modules represent distinctive time features, such that a module-specifically learned time predictor can also accurately decode hourly time from random polling of the same module. These findings open a new paradigm in deciphering the design principle of the biological clock at the system level.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cell Research
生物-细胞生物学
CiteScore
53.90
自引率
0.70%
发文量
2420
审稿时长
2.3 months
期刊介绍:
Cell Research (CR) is an international journal published by Springer Nature in partnership with the Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences (CAS). It focuses on publishing original research articles and reviews in various areas of life sciences, particularly those related to molecular and cell biology. The journal covers a broad range of topics including cell growth, differentiation, and apoptosis; signal transduction; stem cell biology and development; chromatin, epigenetics, and transcription; RNA biology; structural and molecular biology; cancer biology and metabolism; immunity and molecular pathogenesis; molecular and cellular neuroscience; plant molecular and cell biology; and omics, system biology, and synthetic biology. CR is recognized as China's best international journal in life sciences and is part of Springer Nature's prestigious family of Molecular Cell Biology journals.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


