从头设计 pH 响应型自组装螺旋蛋白丝
IF 34.9
1区 材料科学
Q1 MATERIALS SCIENCE, MULTIDISCIPLINARY
引用次数: 0
摘要
生物进化产生了精确、动态的纳米结构,可根据 pH 值和其他环境条件重新配置。然而,设计能对环境做出反应的微米级蛋白质纳米结构仍然是一项挑战。在这里,我们描述了从头设计 pH 响应蛋白质丝的过程,这种蛋白质丝由含有 6 或 9 个埋藏组氨酸残基的亚基构建而成,在中性 pH 条件下可组装成微米级的有序纤维。优化设计的低温电子显微镜结构与计算设计模型的亚基内部几何形状和亚基在纤维中的堆积几乎完全相同。电子显微镜、荧光显微镜和原子力显微镜表征显示,在 0.3 个 pH 单位的范围内,从组装纤维到分解纤维的转变是急剧和可逆的,并且在 pH 值下降后不到 1 秒钟的时间内纤维就会迅速分解。过渡的中点可以通过调节埋藏的含组氨酸氢键网络来调整。因此,计算蛋白质设计为创造能对微小 pH 值变化做出快速反应的非结合纳米材料提供了一条途径。本文章由计算机程序翻译,如有差异,请以英文原文为准。
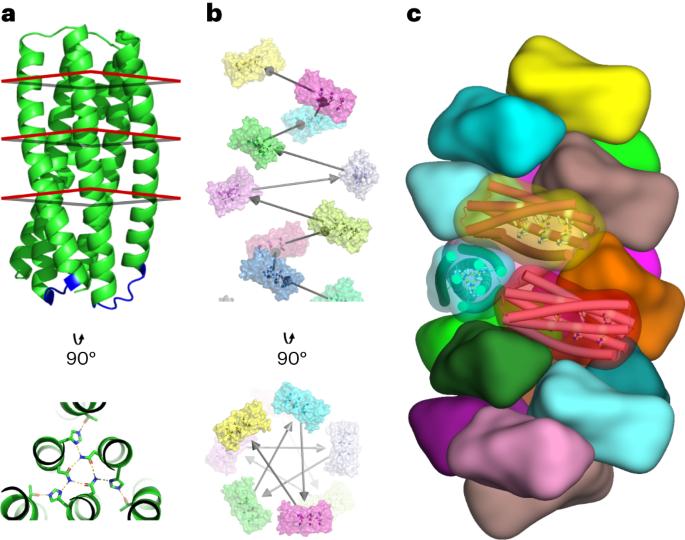
De novo design of pH-responsive self-assembling helical protein filaments
Biological evolution has led to precise and dynamic nanostructures that reconfigure in response to pH and other environmental conditions. However, designing micrometre-scale protein nanostructures that are environmentally responsive remains a challenge. Here we describe the de novo design of pH-responsive protein filaments built from subunits containing six or nine buried histidine residues that assemble into micrometre-scale, well-ordered fibres at neutral pH. The cryogenic electron microscopy structure of an optimized design is nearly identical to the computational design model for both the subunit internal geometry and the subunit packing into the fibre. Electron, fluorescent and atomic force microscopy characterization reveal a sharp and reversible transition from assembled to disassembled fibres over 0.3 pH units, and rapid fibre disassembly in less than 1 s following a drop in pH. The midpoint of the transition can be tuned by modulating buried histidine-containing hydrogen bond networks. Computational protein design thus provides a route to creating unbound nanomaterials that rapidly respond to small pH changes. Engineering the tunability of protein assembly in response to pH changes within a narrow range is challenging. Here the authors report the de novo computational design of pH-responsive protein filaments that exhibit rapid, precise, tunable and reversible assembly and disassembly triggered by small pH changes.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature nanotechnology
工程技术-材料科学:综合
CiteScore
59.70
自引率
0.80%
发文量
196
审稿时长
4-8 weeks
期刊介绍:
Nature Nanotechnology is a prestigious journal that publishes high-quality papers in various areas of nanoscience and nanotechnology. The journal focuses on the design, characterization, and production of structures, devices, and systems that manipulate and control materials at atomic, molecular, and macromolecular scales. It encompasses both bottom-up and top-down approaches, as well as their combinations.
Furthermore, Nature Nanotechnology fosters the exchange of ideas among researchers from diverse disciplines such as chemistry, physics, material science, biomedical research, engineering, and more. It promotes collaboration at the forefront of this multidisciplinary field. The journal covers a wide range of topics, from fundamental research in physics, chemistry, and biology, including computational work and simulations, to the development of innovative devices and technologies for various industrial sectors such as information technology, medicine, manufacturing, high-performance materials, energy, and environmental technologies. It includes coverage of organic, inorganic, and hybrid materials.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


