Lrs14 DNA 结合蛋白家族是 Crenarchaeal Sulfolobales 目中的核相关蛋白
IF 2.6
2区 生物学
Q3 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
古细 胞染色质的组织结构结合了细菌、真核生物的特点。许多古生菌系都蕴藏着种类繁多的小型高表达核团相关蛋白,它们都参与了 DNA 结构的构建。在代表古细菌群模式生物的硫醇杆菌中,Sul7d、Cren7、Sul10a 和 Sul12a 是特征明确的核团相关蛋白。在这里,我们结合了证据,证明 DNA 结合子 Lrs14 家族是硫球菌中核相关蛋白的一部分。Lrs14编码基因广泛存在于不同硫杆菌成员的基因组中,每个基因组通常有4到9个同源物。Lrs14 蛋白含有一个翼螺旋-转螺旋 DNA 结合域,并具有典型的盘绕二聚体。它们具有不同的序列和结构特征,包括氧化还原敏感基团和翻译后修饰的目标残基,从而可将该家族进一步划分为五个保守集群。Lrs14 样蛋白具有独特的 DNA 组织特性。它们以高度合作的方式与 DNA 非序列特异性结合,对富含 AT 的启动子区域略有偏好,从而引入 DNA 扭结,并能对相邻转录单元的转录产生积极或消极的影响。编码 Lrs14 型蛋白的基因在应对各种胁迫条件时表现出相当大的表达差异,某些同源物对特定的胁迫具有特异性。综上所述,我们推测 Lrs14 家族成员可被视为硫杆菌中的核糖体相关蛋白,它们在应激条件下将 DNA 结构作用与全局基因表达作用相结合。本文章由计算机程序翻译,如有差异,请以英文原文为准。
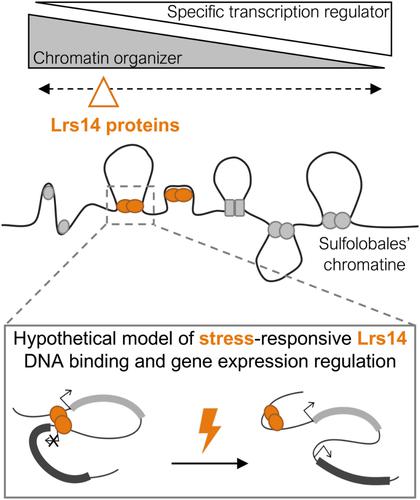
The Lrs14 family of DNA-binding proteins as nucleoid-associated proteins in the Crenarchaeal order Sulfolobales
Organization of archaeal chromatin combines bacterial, eukaryotic, and unique characteristics. Many archaeal lineages harbor a wide diversity of small and highly expressed nucleoid-associated proteins, which are involved in DNA structuring. In Sulfolobales, representing model organisms within the Crenarchaeota, Sul7d, Cren7, Sul10a, and Sul12a are well-characterized nucleoid-associated proteins. Here, we combine evidence that the Lrs14 family of DNA binders is part of the repertoire of nucleoid-associated proteins in Sulfolobales. Lrs14-encoding genes are widespread within genomes of different members of the Sulfolobales, typically encoded as four to nine homologs per genome. The Lrs14 proteins harbor a winged helix-turn-helix DNA-binding domain and are typified by a coiled–coil dimerization. They are characterized by distinct sequence- and structure-based features, including redox-sensitive motifs and residues targeted for posttranslational modification, allowing a further classification of the family into five conserved clusters. Lrs14-like proteins have unique DNA-organizing properties. By binding to the DNA nonsequence specifically and in a highly cooperative manner, with a slight preference for AT-rich promoter regions, they introduce DNA kinks and are able to affect transcription of adjacent transcription units either positively or negatively. Genes encoding Lrs14-type proteins display considerable differential expression themselves in response to various stress conditions, with certain homologs being specific to a particular stressor. Taken together, we postulate that members of the Lrs14 family can be considered nucleoid-associated proteins in Sulfolobales, combining a DNA-structuring role with a global gene expression role in response to stress conditions.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Molecular Microbiology
生物-生化与分子生物学
CiteScore
7.20
自引率
5.60%
发文量
132
审稿时长
1.7 months
期刊介绍:
Molecular Microbiology, the leading primary journal in the microbial sciences, publishes molecular studies of Bacteria, Archaea, eukaryotic microorganisms, and their viruses.
Research papers should lead to a deeper understanding of the molecular principles underlying basic physiological processes or mechanisms. Appropriate topics include gene expression and regulation, pathogenicity and virulence, physiology and metabolism, synthesis of macromolecules (proteins, nucleic acids, lipids, polysaccharides, etc), cell biology and subcellular organization, membrane biogenesis and function, traffic and transport, cell-cell communication and signalling pathways, evolution and gene transfer. Articles focused on host responses (cellular or immunological) to pathogens or on microbial ecology should be directed to our sister journals Cellular Microbiology and Environmental Microbiology, respectively.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


