真核生物和原核生物的共转录基因调控。
IF 81.3
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
摘要
许多 RNA 处理步骤都是在 RNA 聚合酶转录过程中进行的。在原核生物中,共转录活动被认为是司空见惯的事情,因为在原核生物中,由于缺乏膜屏障,从转录到翻译的所有基因表达步骤都可以混合进行。在过去十年中,真核生物中转录和 RNA 处理之间出现了非同寻常的协调。在这篇综述中,我们将讨论我们对真核生物和原核生物共转录基因调控理解的最新进展,比较各种方法和机制,并强调 RNA 聚合酶如何与作用于新生 RNA 的机器相互作用的惊人相似之处。检测瞬时转录和 RNA 处理中间产物的 RNA 测序和成像技术的发展,促进了转录与剪接、3'端裂解和 RNA 动态折叠之间协调的发现,并揭示了处理机制与 RNA 聚合酶之间的物理接触。这些研究表明,内含子在特定新生转录本中的保留可阻止 3'端裂解并导致转录再突破,而这正是真核生物细胞应激反应的标志。我们还讨论了新生 RNA 生物发生和转录之间的协调如何驱动原核生物和真核生物基因表达的基本方面。本文章由计算机程序翻译,如有差异,请以英文原文为准。
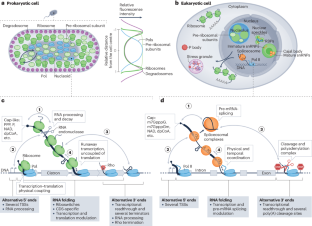
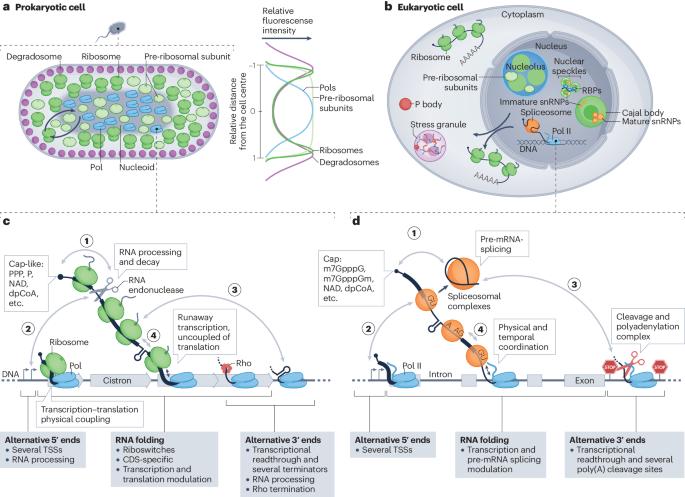
Co-transcriptional gene regulation in eukaryotes and prokaryotes
Many steps of RNA processing occur during transcription by RNA polymerases. Co-transcriptional activities are deemed commonplace in prokaryotes, in which the lack of membrane barriers allows mixing of all gene expression steps, from transcription to translation. In the past decade, an extraordinary level of coordination between transcription and RNA processing has emerged in eukaryotes. In this Review, we discuss recent developments in our understanding of co-transcriptional gene regulation in both eukaryotes and prokaryotes, comparing methodologies and mechanisms, and highlight striking parallels in how RNA polymerases interact with the machineries that act on nascent RNA. The development of RNA sequencing and imaging techniques that detect transient transcription and RNA processing intermediates has facilitated discoveries of transcription coordination with splicing, 3′-end cleavage and dynamic RNA folding and revealed physical contacts between processing machineries and RNA polymerases. Such studies indicate that intron retention in a given nascent transcript can prevent 3′-end cleavage and cause transcriptional readthrough, which is a hallmark of eukaryotic cellular stress responses. We also discuss how coordination between nascent RNA biogenesis and transcription drives fundamental aspects of gene expression in both prokaryotes and eukaryotes. Methodological advances have enabled discoveries of RNA polymerase interactions with RNA processing machineries, such as the splicing and 3′-end cleavage machineries. This Review discusses the roles of these interactions in gene regulation and eukaryotic cellular stress responses, and highlights parallels between co-transcriptional RNA processing in eukaryotes and prokaryotes.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
173.60
自引率
0.50%
发文量
118
审稿时长
6-12 weeks
期刊介绍:
Nature Reviews Molecular Cell Biology is a prestigious journal that aims to be the primary source of reviews and commentaries for the scientific communities it serves. The journal strives to publish articles that are authoritative, accessible, and enriched with easily understandable figures, tables, and other display items. The goal is to provide an unparalleled service to authors, referees, and readers, and the journal works diligently to maximize the usefulness and impact of each article. Nature Reviews Molecular Cell Biology publishes a variety of article types, including Reviews, Perspectives, Comments, and Research Highlights, all of which are relevant to molecular and cell biologists. The journal's broad scope ensures that the articles it publishes reach the widest possible audience.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


