单细胞 NAD(H) 水平可预测克隆淋巴细胞的扩增动态。
IF 17.6
1区 医学
Q1 IMMUNOLOGY
引用次数: 0
摘要
适应性免疫需要高亲和力淋巴细胞从异质库中扩增。目前的模型是通过信号转导来解释这一点的,而我们则假设抗原亲和力会调节离散的代谢途径,从而许可克隆淋巴细胞的动态变化。在这里,我们发现烟酰胺腺嘌呤二核苷酸(NAD)的生物合成是 T 细胞受体亲和力依赖性代谢组的生化枢纽。通过这一核心合成代谢作用,我们发现 NAD 生物合成可控制静止退出检查点,从而为增殖提供节奏。使细胞中的 NAD(H)正常化同样也能使不同亲和力的增殖正常化,而增强 NAD 的生物合成则能使低亲和力克隆扩大。此外,NAD(H)的单细胞差异可以预测T细胞和B细胞在第一次分裂前的分裂潜力,从而消除增殖异质性。我们认为,这支持了一种更广泛的范式,即复杂的信号网络汇聚到代谢途径上,从而控制单细胞的行为。本文章由计算机程序翻译,如有差异,请以英文原文为准。
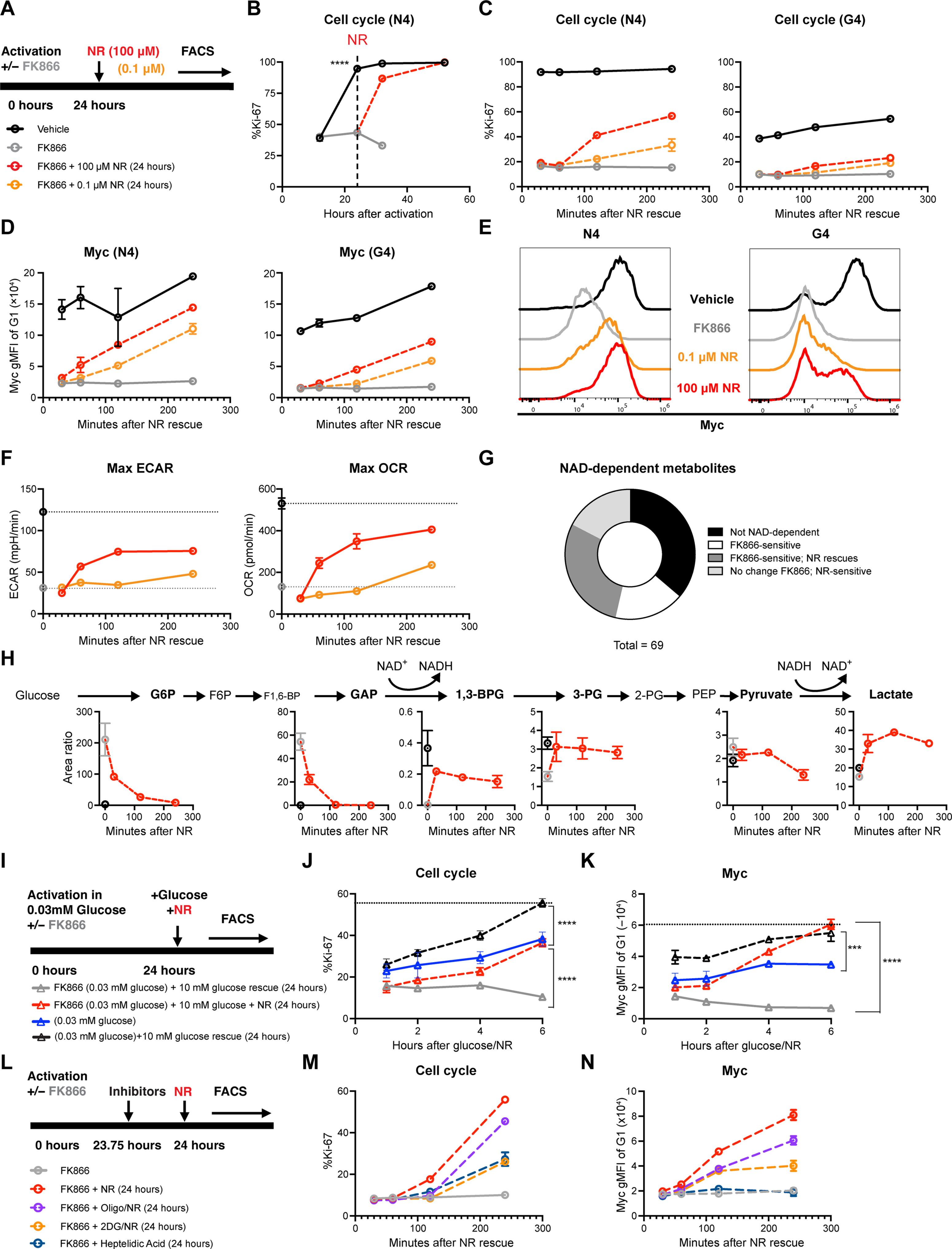
Single-cell NAD(H) levels predict clonal lymphocyte expansion dynamics
Adaptive immunity requires the expansion of high-affinity lymphocytes from a heterogeneous pool. Whereas current models explain this through signal transduction, we hypothesized that antigen affinity tunes discrete metabolic pathways to license clonal lymphocyte dynamics. Here, we identify nicotinamide adenine dinucleotide (NAD) biosynthesis as a biochemical hub for the T cell receptor affinity–dependent metabolome. Through this central anabolic role, we found that NAD biosynthesis governs a quiescence exit checkpoint, thereby pacing proliferation. Normalizing cellular NAD(H) likewise normalizes proliferation across affinities, and enhancing NAD biosynthesis permits the expansion of lower affinity clones. Furthermore, single-cell differences in NAD(H) could predict division potential for both T and B cells, before the first division, unmixing proliferative heterogeneity. We believe that this supports a broader paradigm in which complex signaling networks converge on metabolic pathways to control single-cell behavior.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Science Immunology
Immunology and Microbiology-Immunology
CiteScore
32.90
自引率
2.00%
发文量
183
期刊介绍:
Science Immunology is a peer-reviewed journal that publishes original research articles in the field of immunology. The journal encourages the submission of research findings from all areas of immunology, including studies on innate and adaptive immunity, immune cell development and differentiation, immunogenomics, systems immunology, structural immunology, antigen presentation, immunometabolism, and mucosal immunology. Additionally, the journal covers research on immune contributions to health and disease, such as host defense, inflammation, cancer immunology, autoimmunity, allergy, transplantation, and immunodeficiency. Science Immunology maintains the same high-quality standard as other journals in the Science family and aims to facilitate understanding of the immune system by showcasing innovative advances in immunology research from all organisms and model systems, including humans.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


