短串联重复序列的序列组成变化:异质性、检测、机制和临床意义
IF 52
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
摘要
短串联重复序列(STR)是一类重复元件,由 1-6 个碱基对序列图案的串联阵列组成,占人类基因组的很大一部分。这些疾病的发病年龄、严重程度、渗透性和/或临床表型受重复序列的长度及其序列组成的影响。根据重复序列的类型、频率和位置,非规范基序的存在可通过改变体细胞和代际重复的稳定性、基因表达以及突变转录本介导和/或蛋白质介导的毒性来改变临床结果。在此,我们回顾了重复扩增的各种结构构象、表征序列组成变化的技术进展、其临床相关性以及对疾病机制的影响。本文章由计算机程序翻译,如有差异,请以英文原文为准。
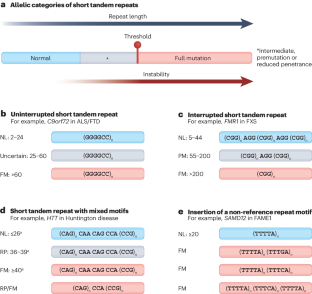
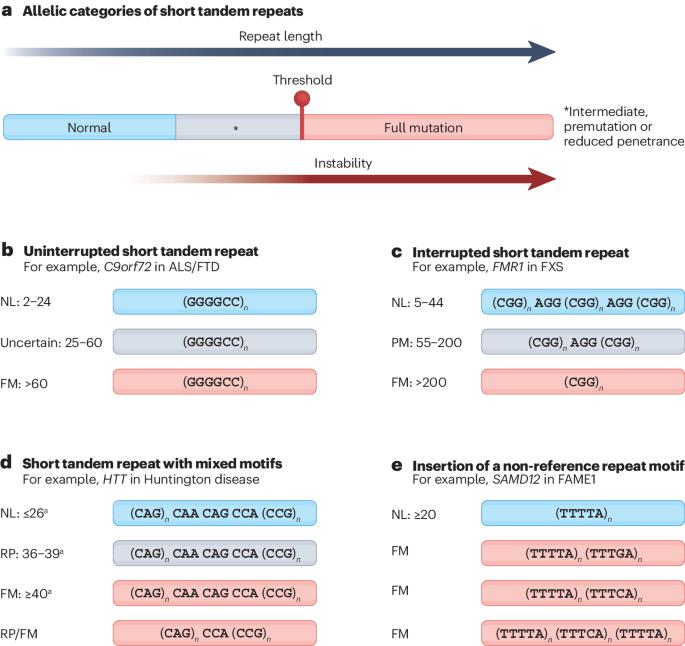
Sequence composition changes in short tandem repeats: heterogeneity, detection, mechanisms and clinical implications
Short tandem repeats (STRs) are a class of repetitive elements, composed of tandem arrays of 1–6 base pair sequence motifs, that comprise a substantial fraction of the human genome. STR expansions can cause a wide range of neurological and neuromuscular conditions, known as repeat expansion disorders, whose age of onset, severity, penetrance and/or clinical phenotype are influenced by the length of the repeats and their sequence composition. The presence of non-canonical motifs, depending on the type, frequency and position within the repeat tract, can alter clinical outcomes by modifying somatic and intergenerational repeat stability, gene expression and mutant transcript-mediated and/or protein-mediated toxicities. Here, we review the diverse structural conformations of repeat expansions, technological advances for the characterization of changes in sequence composition, their clinical correlations and the impact on disease mechanisms. This Review highlights the diversity in sequence composition of disease-related short tandem repeats. The authors discuss how to detect non-canonical motifs in repeat sequences from sequencing data and review the molecular and clinical consequences of sequence composition changes.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Nature Reviews Genetics
生物-遗传学
CiteScore
57.40
自引率
0.50%
发文量
113
审稿时长
6-12 weeks
期刊介绍:
At Nature Reviews Genetics, our goal is to be the leading source of reviews and commentaries for the scientific communities we serve. We are dedicated to publishing authoritative articles that are easily accessible to our readers. We believe in enhancing our articles with clear and understandable figures, tables, and other display items. Our aim is to provide an unparalleled service to authors, referees, and readers, and we are committed to maximizing the usefulness and impact of each article we publish.
Within our journal, we publish a range of content including Research Highlights, Comments, Reviews, and Perspectives that are relevant to geneticists and genomicists. With our broad scope, we ensure that the articles we publish reach the widest possible audience.
As part of the Nature Reviews portfolio of journals, we strive to uphold the high standards and reputation associated with this esteemed collection of publications.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


