同义位点上的选择:不需要的转录本假说。
IF 39.1
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
摘要
虽然在人类中不容易观察到有利于与最丰富的 tRNA 相匹配的密码子的翻译选择,但人类对同义突变还是有选择的。我们假设,这种同义位点选择的大部分原因是为了防止不需要的 RNA(虚假转录本、错误拼接形式或来自可转座元件或病毒的 RNA)。我们认为,同义位点选择的作用不仅在于降低无用转录本的产生率(例如,通过选择外显子剪接增强子和隐性剪接位点),而且在于高GC含量(但低CpG含量)以及内含子的存在和位置既是功能性原生mRNA所特有的,也用于识别原生转录本。最近的研究表明,转录、核输出、液相凝结和 RNA 降解都会促进富含 GC 的转录本,抑制富含 AU/CpG 的转录本,从而支持了这一假设。由于这些 "陷阱 "是针对富含 AU/CpG 的转录本设置的,原生基因的密码子用法反过来也在进化,以避免这种抑制。针对富含 AU/CpG 转录本的平行过滤器也会影响 RNA 的内体导入,这进一步支持了同义位点选择的无用转录本假说,并解释了成功使用转基因和 RNA 疫苗的类似设计规则。本文章由计算机程序翻译,如有差异,请以英文原文为准。
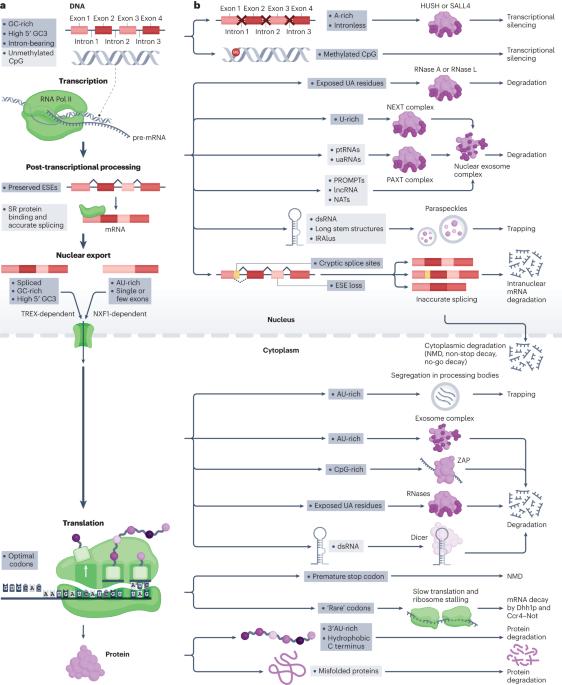
Selection on synonymous sites: the unwanted transcript hypothesis
Although translational selection to favour codons that match the most abundant tRNAs is not readily observed in humans, there is nonetheless selection in humans on synonymous mutations. We hypothesize that much of this synonymous site selection can be explained in terms of protection against unwanted RNAs — spurious transcripts, mis-spliced forms or RNAs derived from transposable elements or viruses. We propose not only that selection on synonymous sites functions to reduce the rate of creation of unwanted transcripts (for example, through selection on exonic splice enhancers and cryptic splice sites) but also that high-GC content (but low-CpG content), together with intron presence and position, is both particular to functional native mRNAs and used to recognize transcripts as native. In support of this hypothesis, transcription, nuclear export, liquid phase condensation and RNA degradation have all recently been shown to promote GC-rich transcripts and suppress AU/CpG-rich ones. With such ‘traps’ being set against AU/CpG-rich transcripts, the codon usage of native genes has, in turn, evolved to avoid such suppression. That parallel filters against AU/CpG-rich transcripts also affect the endosomal import of RNAs further supports the unwanted transcript hypothesis of synonymous site selection and explains the similar design rules that have enabled the successful use of transgenes and RNA vaccines. Multiple mechanisms have evolved to prevent or trap deleterious unwanted transcripts. The unwanted transcript hypothesis proposes that selection at synonymous sites favours mutations that prevent the generation of unwanted transcripts or that make native transcripts appear ‘wanted’ by being GC-rich.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Nature Reviews Genetics
生物-遗传学
CiteScore
57.40
自引率
0.50%
发文量
113
审稿时长
6-12 weeks
期刊介绍:
At Nature Reviews Genetics, our goal is to be the leading source of reviews and commentaries for the scientific communities we serve. We are dedicated to publishing authoritative articles that are easily accessible to our readers. We believe in enhancing our articles with clear and understandable figures, tables, and other display items. Our aim is to provide an unparalleled service to authors, referees, and readers, and we are committed to maximizing the usefulness and impact of each article we publish.
Within our journal, we publish a range of content including Research Highlights, Comments, Reviews, and Perspectives that are relevant to geneticists and genomicists. With our broad scope, we ensure that the articles we publish reach the widest possible audience.
As part of the Nature Reviews portfolio of journals, we strive to uphold the high standards and reputation associated with this esteemed collection of publications.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


