RaPID 对 SARS-CoV-2 的回应
摘要
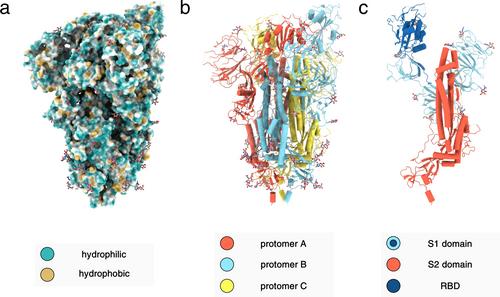
1 引言1.1 RaPID 平台和大环肽药物基因编码肽库已成为新药发现的强大资源。1, 2 这些肽库嵌入了最先进的展示技术,有助于从广阔的序列空间中识别高亲和性肽配体。对肽库进行化学修饰,包括加入非典型氨基酸,可以大大提高筛选出的肽的多样性和药物亲和性。4-6 因此,大多数肽展示都倾向于使用修饰的受限肽,而不是线性肽,7 因为它们具有有益的药物特性、10 大环肽具有多种拓扑结构,11-14 它们的结构特别适合模拟和破坏蛋白质与蛋白质之间的相互作用。15 同样,大环肽固有的刚性增强了它们的靶向亲和力和代谢稳定性、19 然而,大环肽的分子量相对较低,其合成可及性与小分子相似。11, 17 因此,约束肽在小分子和蛋白质疗法之间的 "金发区"(图 1)取得了平衡,6, 20 使其与未来的疗法开发高度相关、22-24 RaPID(随机非标准肽集成发现)平台25(图 2)有助于鉴定选择性和高亲和力结合肽大环26。RaPID 将 FIT 系统(柔性体外翻译)与 mRNA 显示技术巧妙地结合在一起。27 因此,flexizymes(柔性 tRNA-氨基酰化核酶)被用来实现非经典氨基酸的结合。28 通过这种方式,越来越多的非经典氨基酸成为特色,包括 N-甲基、d-、β- 和 γ-氨基酸。虽然各种环化化学反应与 mRNA 显示兼容,1、29、30 但在大多数 RaPID 筛选中,翻译始于氯乙酰化氨基酸,这些氨基酸在与半胱氨酸残基反应后形成硫醚连接。28 由于使用嘌呤霉素将翻译后的多肽与其遗传信息连接起来,因此可以通过测序恢复亲和性筛选富集多肽的信息、29 RaPID 平台已经为越来越多的靶标制备出高亲和性配体,10、26、28、31 因此被认为有能力为几乎任何给定的蛋白质制备大环配体32。SARS-CoV-2的出现以及由此引发的COVID-19大流行使得疫苗、药物和诊断工具的开发迫在眉睫。已确定了一系列与医学相关的病毒靶标37-42 ,其中大多数疫苗和药物相关研究都是针对尖峰糖蛋白(S)38 和主要蛋白酶(Mpro 或 3CLpro)进行的。同源三聚体嵌入病毒包膜,形成了冠状病毒的特有结构47 。它是病毒与宿主细胞融合不可或缺的部分,决定了 SARS-CoV-2 的宿主范围和趋向性48, 49 。在这一过程中,最重要的是受体结合域(RBD),它与 ACE2 受体结合以进入细胞50 。尖峰蛋白作为病毒进入的介质,加上其免疫原性,使尖峰蛋白成为成功疫苗开发活动中的一个常见元素。
Genetically encoded peptide libraries are at the forefront of de novo drug discovery. The RaPID (Random Nonstandard Peptides Integrated Discovery) platform stands out due to the unique combination of flexible in vitro translation (FIT) and mRNA display. This enables the incorporation of non-canonical amino acids, improving chemical diversity and allowing macrocyclisation of the peptide library. The resulting constrained peptides are valued for their strong binding affinity and stability, especially in the context of protein-protein interactions. In response to SARS-CoV-2, the causative agent of the COVID-19 pandemic, the RaPID system proved valuable in identifying high-affinity ligands of viral proteins. Among many peptide ligands of SARS-CoV-2 spike and main protease (Mpro), several macrocycles stand out for their exceptional binding affinities. Structural data showcases distinct binding modes in complex with the receptor-binding domain (RBD) of the spike glycoprotein or the catalytic active site of Mpro. However, translating these in vitro findings into clinical applications remains challenging, especially due to insufficient cell permeability.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


