塑造 rRNA 和 tRNA 基因的染色质景观,RNA 聚合酶 II 转录的新角色?
IF 2.6
4区 生物学
Q4 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
真核生物基因必须凝集到染色质中,同时转录机制仍可利用它来支持基因表达。在真核生物的三种 RNA 聚合酶(RNAP)中,RNAPII 是独一无二的,部分原因在于其最大亚基 Rpb1 的 C 端结构域(CTD)。在转录周期中,Rpb1 CTD 可被广泛修饰,从而在共转录过程中招募特定的相互作用蛋白。其中包括控制染色质开放或关闭的染色质重塑因子。人们对无 CTD 的 RNAPI 和 RNAPIII 如何处理 rRNA 和 tRNA 基因上的染色质了解较少。在此,我们回顾了我们对 tRNA 基因和 rRNA 基因的染色质如何重塑以响应酵母环境线索的理解的最新进展,尤其关注局部 RNAPII 转录在这些位点招募染色质重塑因子的作用。在裂殖酵母中,tRNA 基因上的 RNAPII 转录对于重建允许 tRNA 转录的染色质环境非常重要,这有助于从静止期开始的生长。与此相反,rRNA 基因上的局部 RNAPII 转录与芽殖酵母和裂殖酵母饥饿时染色质的关闭有关,这表明 RNAPII 在建立沉默染色质方面发挥作用。这些相反的作用可能支持一种普遍的模式,即 RNAPII 转录将染色质重塑者招募到 tRNA 和 rRNA 基因上,以促进染色质的关闭和重新开放,从而对环境做出反应。本文章由计算机程序翻译,如有差异,请以英文原文为准。
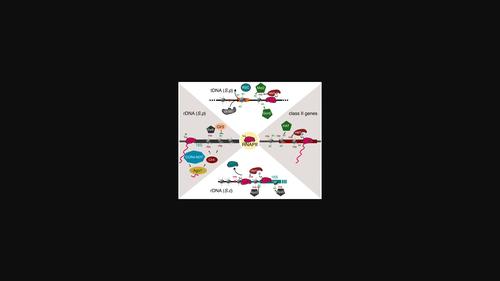
Shaping the chromatin landscape at rRNA and tRNA genes, an emerging new role for RNA polymerase II transcription?
Eukaryotic genes must be condensed into chromatin while remaining accessible to the transcriptional machinery to support gene expression. Among the three eukaryotic RNA polymerases (RNAP), RNAPII is unique, partly because of the C-terminal domain (CTD) of its largest subunit, Rpb1. Rpb1 CTD can be extensively modified during the transcription cycle, allowing for the co-transcriptional recruitment of specific interacting proteins. These include chromatin remodeling factors that control the opening or closing of chromatin. How the CTD-less RNAPI and RNAPIII deal with chromatin at rRNA and tRNA genes is less understood. Here, we review recent advances in our understanding of how the chromatin at tRNA genes and rRNA genes can be remodeled in response to environmental cues in yeast, with a particular focus on the role of local RNAPII transcription in recruiting chromatin remodelers at these loci. In fission yeast, RNAPII transcription at tRNA genes is important to re-establish a chromatin environment permissive to tRNA transcription, which supports growth from stationary phase. In contrast, local RNAPII transcription at rRNA genes correlates with the closing of the chromatin in starvation in budding and fission yeast, suggesting a role in establishing silent chromatin. These opposite roles might support a general model where RNAPII transcription recruits chromatin remodelers to tRNA and rRNA genes to promote the closing and reopening of chromatin in response to the environment.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Yeast
生物-生化与分子生物学
CiteScore
5.30
自引率
3.80%
发文量
55
审稿时长
3 months
期刊介绍:
Yeast publishes original articles and reviews on the most significant developments of research with unicellular fungi, including innovative methods of broad applicability. It is essential reading for those wishing to keep up to date with this rapidly moving field of yeast biology.
Topics covered include: biochemistry and molecular biology; biodiversity and taxonomy; biotechnology; cell and developmental biology; ecology and evolution; genetics and genomics; metabolism and physiology; pathobiology; synthetic and systems biology; tools and resources
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


