人类微生物组的系统挖掘识别具有不同活性谱的抗菌肽。
IF 20.5
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
摘要
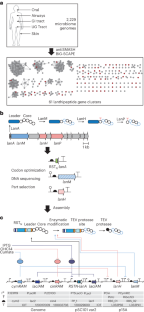
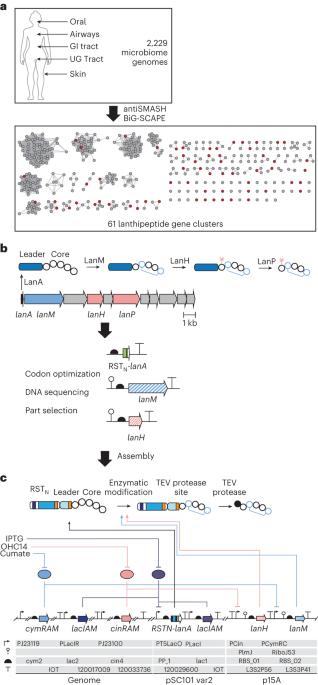
人类相关的细菌分泌修饰肽来控制宿主生理和重塑微生物群的物种组成。在这里,我们扫描了2229个人类微生物组计划物种的基因组,这些物种定植于皮肤、胃肠道、泌尿生殖道、口腔和气管,以寻找编码RiPPs(核糖体合成和翻译后修饰肽)的基因簇。共获得218个lanthipeptide和25个lasso peptide,其中70个在大肠杆菌中合成并表达,23个可以纯化并进行功能表征。测试了它们对与健康人类菌群和病原体相关的细菌的活性。发现了新的抗生素,用于对抗与皮肤、鼻腔和阴道生态失调有关的菌株,以及选择性靶向肠道菌群的口服菌株。广谱和窄谱抗生素可用于耐甲氧西林金黄色葡萄球菌和耐万古霉素肠球菌。挖掘与人类相关的微生物产生的天然产物将有助于阐明生态关系,并可能成为抗菌药物发现的丰富资源。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Systematic mining of the human microbiome identifies antimicrobial peptides with diverse activity spectra
Human-associated bacteria secrete modified peptides to control host physiology and remodel the microbiota species composition. Here we scanned 2,229 Human Microbiome Project genomes of species colonizing skin, gastrointestinal tract, urogenital tract, mouth and trachea for gene clusters encoding RiPPs (ribosomally synthesized and post-translationally modified peptides). We found 218 lanthipeptides and 25 lasso peptides, 70 of which were synthesized and expressed in E. coli and 23 could be purified and functionally characterized. They were tested for activity against bacteria associated with healthy human flora and pathogens. New antibiotics were identified against strains implicated in skin, nasal and vaginal dysbiosis as well as from oral strains selectively targeting those in the gut. Extended- and narrow-spectrum antibiotics were found against methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococci. Mining natural products produced by human-associated microbes will enable the elucidation of ecological relationships and may be a rich resource for antimicrobial discovery. An analysis of over 2,000 genomes from Human Microbiome Project metagenomic data led to the identification of several extended- and narrow-spectrum antibiotics against clinical multidrug-resistant pathogens, including methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococci.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


