专性生物营养霜霉在拟南芥中持续诱导几乎相同的保护性微生物组。
IF 20.5
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
摘要
拟南芥透明operonospora Arabidopsis (Hpa)是一种专性生物营养霜霉病,通常在含有复杂微生物群的拟南芥宿主上培养。我们假设,除了病原体外,培养过程还会增殖Hpa相关微生物群(HAM),并利用该模型系统研究哪些微生物与Hpa始终相关。利用扩增子测序,我们发现9个细菌序列变异在荷兰和德国的4个Hpa培养物中至少有3个共有,占受感染植物层圈群落的34%。全基因组测序显示,从这些不同的Hpa培养物中分离出的具有代表性的HAM细菌是等基因的,另外七个已发表的Hpa宏基因组包含了许多HAM序列。虽然我们发现HAM从Hpa感染中受益,但HAM对Hpa孢子的形成有负面影响。此外,我们发现被病原体感染的植物可以选择性地将HAM招募到它们的根和芽中,并形成一个与土壤传播的感染相关的微生物组,帮助抵抗病原体。了解感染相关微生物群形成的机制可能有助于培育选择保护性微生物群的作物品种。本文章由计算机程序翻译,如有差异,请以英文原文为准。
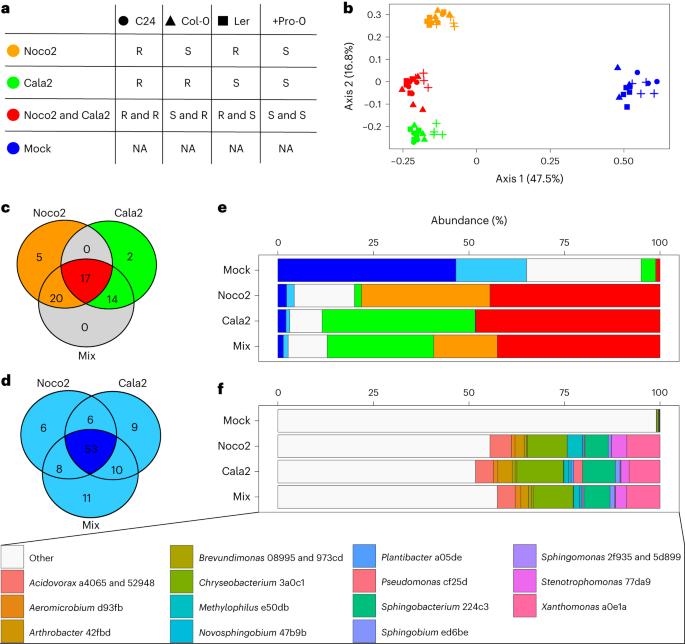
Obligate biotroph downy mildew consistently induces near-identical protective microbiomes in Arabidopsis thaliana
Hyaloperonospora arabidopsidis (Hpa) is an obligately biotrophic downy mildew that is routinely cultured on Arabidopsis thaliana hosts that harbour complex microbiomes. We hypothesized that the culturing procedure proliferates Hpa-associated microbiota (HAM) in addition to the pathogen and exploited this model system to investigate which microorganisms consistently associate with Hpa. Using amplicon sequencing, we found nine bacterial sequence variants that are shared between at least three out of four Hpa cultures in the Netherlands and Germany and comprise 34% of the phyllosphere community of the infected plants. Whole-genome sequencing showed that representative HAM bacterial isolates from these distinct Hpa cultures are isogenic and that an additional seven published Hpa metagenomes contain numerous sequences of the HAM. Although we showed that HAM benefit from Hpa infection, HAM negatively affect Hpa spore formation. Moreover, we show that pathogen-infected plants can selectively recruit HAM to both their roots and shoots and form a soil-borne infection-associated microbiome that helps resist the pathogen. Understanding the mechanisms by which infection-associated microbiomes are formed might enable breeding of crop varieties that select for protective microbiomes. Near-identical downy-mildew-associated microbiomes are recruited by infected Arabidopsis plants in different laboratories and reduce the impact of subsequent infection.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


