强迫症外显子组测序揭示了罕见损伤性编码变异的负担
IF 21.2
1区 医学
Q1 NEUROSCIENCES
引用次数: 24
摘要
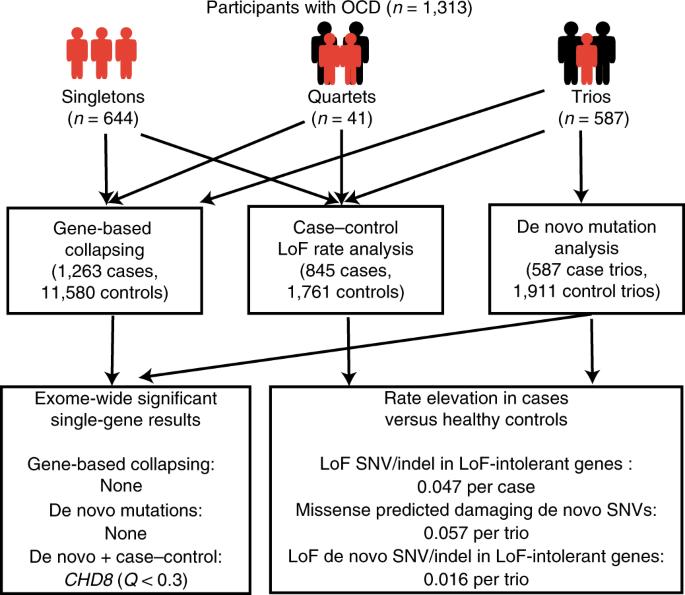
强迫症(OCD)影响着1-2%的人口,与其他复杂的神经精神疾病一样,人们认为罕见变异也会导致其遗传风险。在这项研究中,我们对迄今为止最大的强迫症队列(总病例数为 1313 例,包括 587 个三联病例、41 个四联病例和 644 个单联病例)进行了外显子组测序,并描述了罕见损伤性编码变异对疾病风险的贡献。在病例对照分析(n = 1,263/11,580)中,SLITRK5 的单基因结果最为显著(比值比 (OR) = 8.8,95% 置信区间为 3.4-22.5,P = 2.3 × 10-6)。在整个外显子组中,功能缺失(LoF)变异过多,特别是在不耐受 LoF 的基因中(OR = 1.33,P = 0.01)。在对三组基因的分析中,我们观察到相对于对照组,有过量的从头错义预测损伤性变异(OR = 1.22,P = 0.02),同时在不耐受 LoF 的基因中也有过量的从头 LoF 突变(OR = 2.55,P = 7.33 × 10-3)。这些数据支持罕见编码变异对强迫症遗传风险的贡献。对迄今为止最大的强迫症患者外显子组测序数据集(n = 1,313 例患者)的分析显示,病例对照研究和从头变异研究都支持罕见损伤性编码变异对强迫症遗传风险的贡献。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Exome sequencing in obsessive–compulsive disorder reveals a burden of rare damaging coding variants
Obsessive–compulsive disorder (OCD) affects 1–2% of the population, and, as with other complex neuropsychiatric disorders, it is thought that rare variation contributes to its genetic risk. In this study, we performed exome sequencing in the largest OCD cohort to date (1,313 total cases, consisting of 587 trios, 41 quartets and 644 singletons of affected individuals) and describe contributions to disease risk from rare damaging coding variants. In case–control analyses (n = 1,263/11,580), the most significant single-gene result was observed in SLITRK5 (odds ratio (OR) = 8.8, 95% confidence interval 3.4–22.5, P = 2.3 × 10−6). Across the exome, there was an excess of loss of function (LoF) variation specifically within genes that are LoF-intolerant (OR = 1.33, P = 0.01). In an analysis of trios, we observed an excess of de novo missense predicted damaging variants relative to controls (OR = 1.22, P = 0.02), alongside an excess of de novo LoF mutations in LoF-intolerant genes (OR = 2.55, P = 7.33 × 10−3). These data support a contribution of rare coding variants to OCD genetic risk. An analysis of the largest exome sequencing dataset of people with obsessive–compulsive disorder to date (n = 1,313 affected individuals), where both case–control and de novo variant studies support a contribution of rare damaging coding variants to risk.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature neuroscience
医学-神经科学
CiteScore
38.60
自引率
1.20%
发文量
212
审稿时长
1 months
期刊介绍:
Nature Neuroscience, a multidisciplinary journal, publishes papers of the utmost quality and significance across all realms of neuroscience. The editors welcome contributions spanning molecular, cellular, systems, and cognitive neuroscience, along with psychophysics, computational modeling, and nervous system disorders. While no area is off-limits, studies offering fundamental insights into nervous system function receive priority.
The journal offers high visibility to both readers and authors, fostering interdisciplinary communication and accessibility to a broad audience. It maintains high standards of copy editing and production, rigorous peer review, rapid publication, and operates independently from academic societies and other vested interests.
In addition to primary research, Nature Neuroscience features news and views, reviews, editorials, commentaries, perspectives, book reviews, and correspondence, aiming to serve as the voice of the global neuroscience community.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


