分析多尺度三维基因组组织的计算方法。
IF 39.1
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
摘要
全基因组图谱和成像技术的最新进展使基因组在细胞核中的空间组织和折叠得以表征。与此同时,已经开发了先进的计算方法来利用这些映射数据来揭示多尺度三维(3D)基因组特征,并提供基因组结构及其与转录等基因组功能的联系的更完整的视图。在这里,我们讨论了最近开发的计算工具,包括基于机器学习的方法和综合结构建模框架,是如何系统地、多尺度地描述3D基因组组织、基因组和表观基因组特征、功能核成分和基因组功能的不同尺度之间的联系的。然而,仍然需要更全面地整合各种基因组和成像数据集的方法来揭示3D基因组结构在定义健康和疾病中的细胞表型中的功能作用。本文章由计算机程序翻译,如有差异,请以英文原文为准。
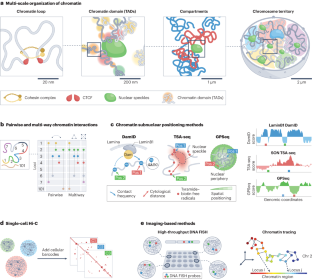
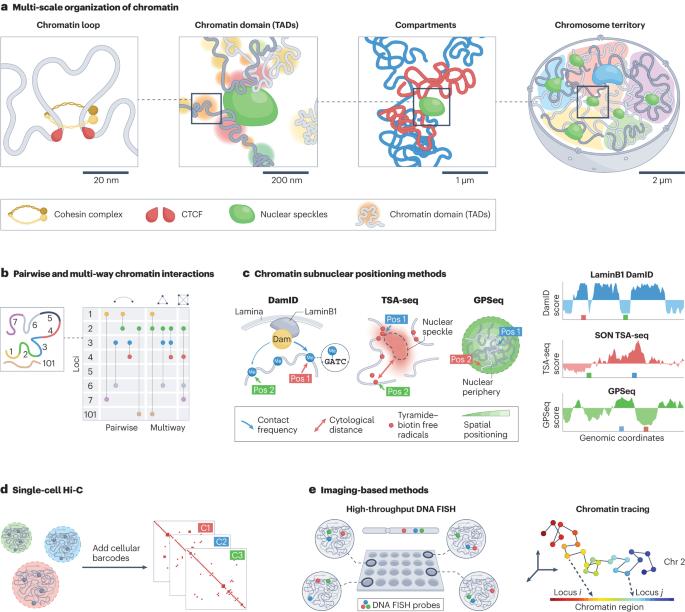
Computational methods for analysing multiscale 3D genome organization
Recent progress in whole-genome mapping and imaging technologies has enabled the characterization of the spatial organization and folding of the genome in the nucleus. In parallel, advanced computational methods have been developed to leverage these mapping data to reveal multiscale three-dimensional (3D) genome features and to provide a more complete view of genome structure and its connections to genome functions such as transcription. Here, we discuss how recently developed computational tools, including machine-learning-based methods and integrative structure-modelling frameworks, have led to a systematic, multiscale delineation of the connections among different scales of 3D genome organization, genomic and epigenomic features, functional nuclear components and genome function. However, approaches that more comprehensively integrate a wide variety of genomic and imaging datasets are still needed to uncover the functional role of 3D genome structure in defining cellular phenotypes in health and disease. In this Review, Zhang et al. discuss how recent advances in computational methods are helping to reveal the multiscale features involved in genome folding within the nucleus and how the resulting 3D genome organization relates to genome function.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Nature Reviews Genetics
生物-遗传学
CiteScore
57.40
自引率
0.50%
发文量
113
审稿时长
6-12 weeks
期刊介绍:
At Nature Reviews Genetics, our goal is to be the leading source of reviews and commentaries for the scientific communities we serve. We are dedicated to publishing authoritative articles that are easily accessible to our readers. We believe in enhancing our articles with clear and understandable figures, tables, and other display items. Our aim is to provide an unparalleled service to authors, referees, and readers, and we are committed to maximizing the usefulness and impact of each article we publish.
Within our journal, we publish a range of content including Research Highlights, Comments, Reviews, and Perspectives that are relevant to geneticists and genomicists. With our broad scope, we ensure that the articles we publish reach the widest possible audience.
As part of the Nature Reviews portfolio of journals, we strive to uphold the high standards and reputation associated with this esteemed collection of publications.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


