环境DNA分类分配的系统发育定位解释
IF 2.1
4区 生物学
Q3 MICROBIOLOGY
引用次数: 0
摘要
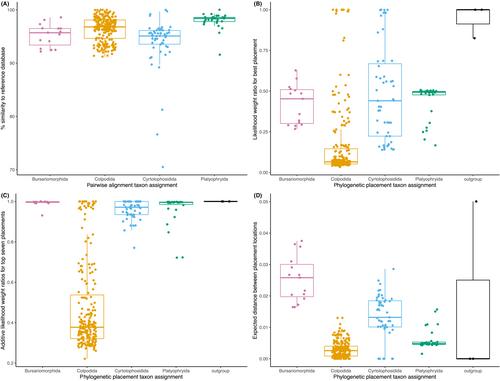
操作分类单元(OTUs)的分类分配是分析环境测序数据的重要生物信息学步骤。配对比对和系统发育定位方法代表了两种可选的分类分配方法,但它们的结果可能不同。在这里,我们利用森林土壤中可用的阴道目纤毛虫OTUs来比较VSEARCH(进行配对比对)和EPA-ng(进行系统发育定位)的分类定位。结果表明,在亚分类单元水平上,当配对比对和系统发育定位之间的分类归属存在差异时,otu与参考数据库的配对相似性较低。然后,我们展示了如何使用GAPPA进一步评估EPA-ng的输出,当存在多个等可能的OTU位置时,通过考虑子分类单元中OTU位置的似然权重之和以及等可能位置之间的分支距离来评估分类分配。我们还利用它们在亚类群中的位置推断了阴道目OTUs的进化和生态特征。本研究演示了如何通过使用GAPPA结合感兴趣的进化枝的分类多样性知识来全面分析EPA-ng的输出。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Interpreting phylogenetic placements for taxonomic assignment of environmental DNA
Taxonomic assignment of operational taxonomic units (OTUs) is an important bioinformatics step in analyzing environmental sequencing data. Pairwise alignment and phylogenetic‐placement methods represent two alternative approaches to taxonomic assignments, but their results can differ. Here we used available colpodean ciliate OTUs from forest soils to compare the taxonomic assignments of VSEARCH (which performs pairwise alignments) and EPA‐ng (which performs phylogenetic placements). We showed that when there are differences in taxonomic assignments between pairwise alignments and phylogenetic placements at the subtaxon level, there is a low pairwise similarity of the OTUs to the reference database. We then showcase how the output of EPA‐ng can be further evaluated using GAPPA to assess the taxonomic assignments when there exist multiple equally likely placements of an OTU, by taking into account the sum over the likelihood weights of the OTU placements within a subtaxon, and the branch distances between equally likely placement locations. We also inferred the evolutionary and ecological characteristics of the colpodean OTUs using their placements within subtaxa. This study demonstrates how to fully analyze the output of EPA‐ng, by using GAPPA in conjunction with knowledge of the taxonomic diversity of the clade of interest.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
4.30
自引率
4.50%
发文量
85
审稿时长
6-12 weeks
期刊介绍:
The Journal of Eukaryotic Microbiology publishes original research on protists, including lower algae and fungi. Articles are published covering all aspects of these organisms, including their behavior, biochemistry, cell biology, chemotherapy, development, ecology, evolution, genetics, molecular biology, morphogenetics, parasitology, systematics, and ultrastructure.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


